Figure 2.
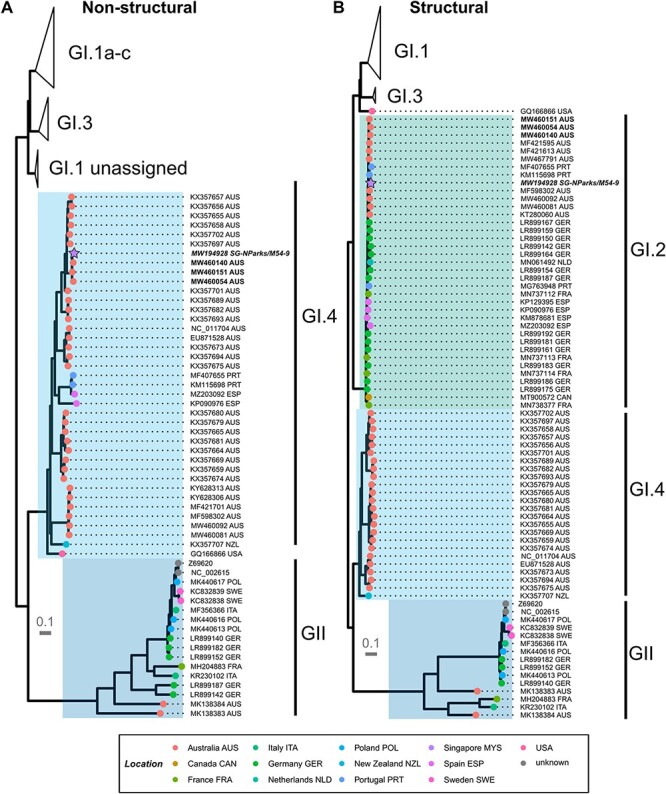
Maximum-likelihood phylogenies of representative global lagovirus sequences. Phylogenies were estimated in IQTREE2 using the best-fit model as selected by ModelFinder and were estimated separately for the (A) NS and (B) S sequences. The GI.4cP-GI.2 viruses are identified in bold, while the Singapore RHDV 2020 sequence (NCBI accession number MW194928) is presented in bold italics. Branch support was estimated with 1,000 ultrafast bootstrap replicates and 1,000 replicates of the SH-aLRT test. The scale bar shows nucleotide substitutions per site. Tips were coloured based on the location from which the sequence was reported. Highlighted regions show (from bottom to top) genogroup GII , genotype GI.4 , genotype GI.2 , genotype GI.1 , and genotype GI.3 . One sequence is unclassified in the structural regions and has not been highlighted. GI.1 and GI.3 clades have been collapsed.
