ABSTRACT
Dissemination of hypervirulent and carbapenem-resistant Klebsiella pneumoniae (CRKP) has been reported worldwide, posing a serious threat to antimicrobial therapy and public health. Outer membrane vesicles (OMVs) act as vectors for the horizontal transfer of virulence and resistance genes. However, K. pneumoniae OMVs that transfer carbapenem resistance genes into hypervirulent K. pneumoniae (hvKP) have been insufficiently investigated. Therefore, this study investigates the transmission of the blaNDM-1 gene encoding resistance via OMVs released from CRKP and the potential mechanism responsible for the carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) emergence. OMVs were isolated via ultracentrifugation from CRKP with or without meropenem selective pressure. OMVs were then used to transform classical K. pneumoniae (ckp) ATCC 10031, extended-spectrum β-lactamase (ESBL)-producing K. pneumoniae ATCC 700603, and hvKP NTUH-K2044. Our results showed that meropenem treatment resulted in changes in the number and diameter of OMVs secreted by CRKP. OMVs derived from CRKP mediated the transfer of blaNDM-1 to ckp and hvKP, thereby increasing the carbapenem MIC of transformants. Further experiments confirmed that NTUH-K2044 transformants exhibited hypervirulence. Our study demonstrates, for the first time, that OMVs derived from CRKP can carry blaNDM-1 and deliver resistance genes to other K. pneumoniae strains, even hvKP. The transfer of carbapenem genes into hypervirulent strains may promote the emergence and dissemination of CR-hvKP. This study elucidates a new mechanism underlying the formation of CR-hvKP.
KEYWORDS: outer membrane vesicles, carbapenem-resistant Klebsiella pneumoniae, horizontal gene transfer, bla NDM-1 , carbapenem-resistant hypervirulent Klebsiella pneumoniae
INTRODUCTION
Klebsiella pneumoniae is a major opportunistic pathogen that causes community-acquired and hospital-acquired infections. K. pneumoniae can acquire exogenous resistance- and virulence-encoding genetic elements. Carbapenem-resistant K. pneumoniae (CRKP) has been spreading rapidly worldwide, posing a serious challenge to antimicrobial therapy (1). Hypervirulent K. pneumoniae (hvKP) can cause community-acquired infections and severe invasive infections, even in community-dwelling healthy hosts, which can result in septic liver abscesses, endophthalmitis, and meningitis (2).
The generation of K. pneumoniae strains that are hypervirulent (hv) and carbapenem resistant poses a serious threat to public health (3) and has three different evolutionary paths: hvKP strains may acquire drug-resistant plasmids, forming carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) (4–7); CRKP strains may acquire virulence plasmids, forming hv-CRKP (8); and virulence and carbapenem resistance genetic elements may integrate into a single plasmid (9). In hvKP, horizontal gene transfer (HGT) is constrained by the substantially thickened polysaccharide capsule and the limited chromosomal recombination; the reduced plasmid diversity results in a reduction in pangenomic diversity (10). These factors mean that CRKP clones acquire virulence plasmids more efficiently than hvKP clones acquire drug-resistant plasmids. Although difficult, hvKP can acquire drug-resistant plasmids and generate CR-hvKP. Developing a better understanding of the mechanisms contributing to the transmission of drug-resistant plasmids that promote the emergence of CR-hvKP is critical.
HGT is an important mechanism leading to the transfer of drug resistance and virulence genes. Currently, the most widely described HGT mechanisms are conjugation, transformation, and transduction (11–13). Although these mechanisms conduce to gene transfer within bacteria, they have limitations, including dependence on cell-to-cell contact, bacterial competence, and host specificity. Recent studies have reported a new mechanism of outer membrane vesicles (OMVs) mediating HGT.
OMVs are 20- to 250-nm spherical bilayer liposomes, naturally released by Gram-negative bacteria during growth. OMVs originate from the outer membrane and enclose lipopolysaccharide (LPS), phospholipids, genetic elements (DNA, RNA, and plasmids), and periplasmic as well as cytoplasmic protein components (14). As part of a secretion-delivery system, OMVs allow long-distance dissemination of bacterial components to the environment and facilitate interactions between cells without the need for direct contact. The components within the lumen of vesicles are protected from DNase, making OMVs an effective HGT vector (15).
Since 1999, studies have reported that OMVs can carry virulence and resistance genes that can be horizontally transferred to bacteria (16). Dell’Annunziata et al. (17) discovered that genetically engineered KP-OMVs could transfer drug-resistant plasmids horizontally and that the transfer frequency correlated with the plasmid copy number. Subsequently, Hua et al. (10) showed that hvKP-OMVs mediated the transmission of virulence genes to extended-spectrum β-lactamase (ESBL)-producing classical K. pneumoniae (ckp). However, it is uncertain whether OMVs derived from CRKP can mediate the dissemination of drug resistance genes to hvKP, thus producing CR-hvKP. In this study, we aimed to investigate the transmission of the blaNDM-1 gene via OMVs released from CRKP and the potential mechanism responsible for the emergence of CR-hvKP.
RESULTS
Characterization of OMVs derived from CRKP under different conditions.
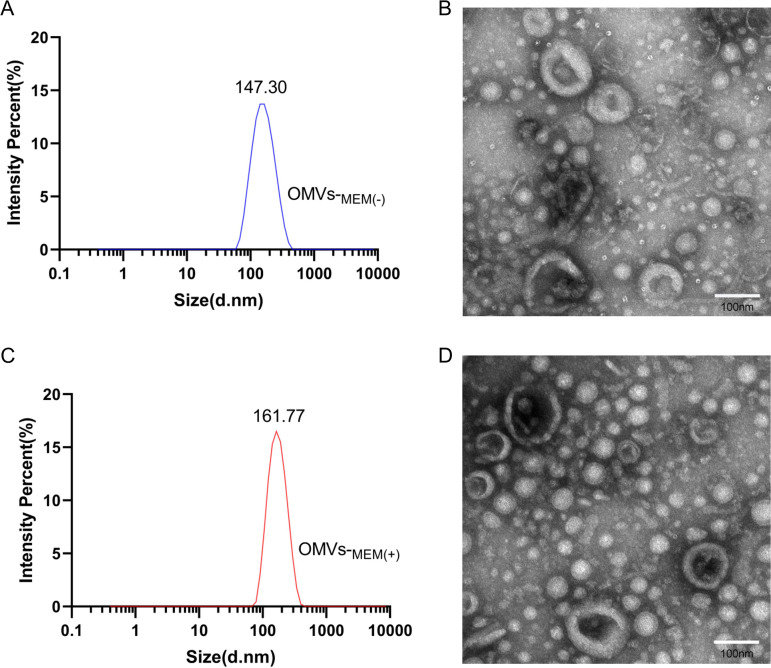
OMVs were extracted and purified from CRKP by ultracentrifugation. Transmission electron microscopy (TEM) showed that the purified OMVs were spherical. There were no substantial morphological differences between OMVs-MEM(−) (Fig. 1B) and OMVs-MEM(+) (Fig. 1D). The dynamic light scattering (DLS) analysis showed that OMVs-MEM(+) were larger (78.8 to 396 nm, median size of 161.77 nm) (Fig. 1C) than OMVs-MEM(−) (68.1 to 396 nm, median size of 147.30 nm) (Fig. 1A). No bacteria were visualized under the microscope, and the contamination controls on the culture plates did not show any growth, thus indicating that the OMVs were well purified.
FIG 1.
Purification and characterization of OMVs derived from CRKP under different conditions. (A and C) DLS of OMVs produced in LB broth without 8 μg/mL meropenem [OMVs-MEM(−)] (A) and OMVs produced in LB broth with 8 μg/mL meropenem [OMVs-MEM(+)] (n = 3) (C). (B and D) TEM of OMVs-MEM(−) (B) and (D) OMVs-MEM(+) (D). Bar, 100 nm.
Protein and DNA packaging in the OMVs.
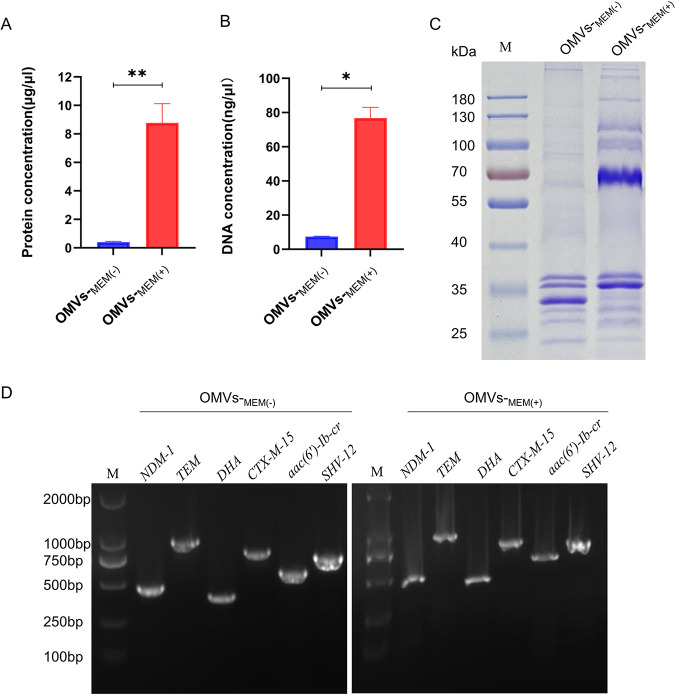
The mean protein concentrations (based on the averages from the three experiments) of OMVs-MEM(−) and OMVs-MEM(+) were 0.405 ± 0.045 μg/μL and 8.777 ± 1.346 μg/μL, respectively (Fig. 2A). The mean DNA concentrations (based on the average from the three experiments) of OMVs-MEM(−) and OMVs-MEM(+) were 7.49 ± 0.14 ng/μL and 76.80 ± 6.21 ng/μL, respectively (Fig. 2B). The DNA purified from OMVs-MEM(−) and OMVs-MEM(+) gave specific amplified products for blaNDM-1, blaCTX-M-15, blaSHV-12, blaTEM-1, blaDHA-1, and aac(6′)-Ib-cr (Fig. 2D). SDS-PAGE was performed on purified OMVs to compare the differences in the protein composition and expression levels in OMVs-MEM(−) and OMVs-MEM(+). The OMVs-MEM(−) and OMVs-MEM(+) protein profiles showed differences in both protein composition and expression levels. Substantially elevated levels of a protein band at approximately 70 kDa were observed in OMVs-MEM(+) compared with that of OMVs-MEM(−) (Fig. 2C).
FIG 2.
Comparative analysis of OMVs under different conditions. (A) Protein concentration of OMVs produced in LB broth without or with the pressure of 8 μg/mL meropenem [OMVs-MEM(−) and OMVs-MEM(+), respectively] (n = 3). (B) DNA concentration of OMVs-MEM(−) and OMVs-MEM(+) (n = 3). (C) Protein profiles presented using gel electrophoresis of OMVs-MEM(−) and OMVs-MEM(+). (D) Detection of resistance genes in OMVs-MEM(−) and OMVs-MEM(+) using PCR. *, P < 0.05, and **, P < 0.01.
OMVs from CRKP can mediate the transfer of blaNDM-1 to ckp and hvKP.
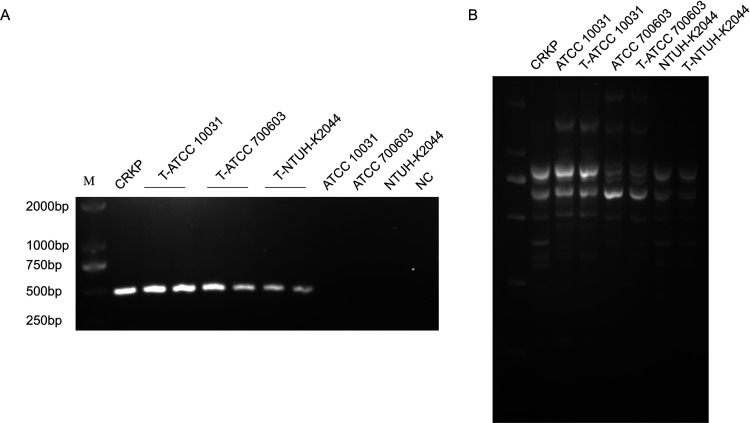
ckp ATCC 10031, ckp ATCC 700603, and hvKP NTUH-K2044 were used as recipient strains and were coincubated with OMVs isolated from CRKP. DNA transformation was first ruled out because no colonies grew on the MacConkey (MAC) plate with 1 μg/mL meropenem when the plasmid was incubated with ckp and hvKP. Additionally, the group of lysed OMVs showed no colonies on the MAC plate with 1 μg/mL meropenem, and only intact OMVs led to meropenem-resistant colony growth. Thus, we concluded that only intact OMVs could mediate the transfer of blaNDM-1. The blaNDM-1 transfer frequencies from 500 μg OMVs-MEM(+) to ckp ATCC 10031 and hvKP NTUH-K2044 were 9.79 × 10−9 ± 3.52 × 10−9 and 7.83 × 10−9 ± 3.48 × 10−9, respectively. However, the transfer frequency from 200 μg OMVs-MEM(+) to ATCC 700603 was much higher (8.08 × 10−7 ± 1.89 × 10−7). When ATCC 700603 was coincubated with 20 μg OMVs, the transfer frequency of the OMVs-MEM(−) group (1.42 × 10−6 ± 0.27 × 10−6) was higher than that of the OMVs-MEM(+) group (1.65 × 10−7 ± 0.25 × 10−7) (Table 1). Two randomly selected transformants from each of the three recipient strain groups were confirmed for blaNDM-1 using PCR and were visualized using agarose gel electrophoresis. All strains were blaNDM-1-positive strains (Fig. 3A). However, all strains were negative for blaCTX-M-15, blaSHV-12, blaTEM-1C, blaDHA-1, and aac(6′)-Ib-cr. To completely understand the results, the whole genome of the NTUH-K2044 transformants was sequenced (GenBank accession number SAMN33420516). The results showed that the plasmid IncFIBpKPHS1 harboring blaNDM-1 was transferred to NTUH-K2044 via OMVs, and blaNDM-1 was the only resistance gene transferred.
TABLE 1.
Frequencies of blaNDM-1 transfer via OMVs from CRKP into recipient bacteria
| Treatment | Amt of OMVs (μg) | Transfer frequency |
|---|---|---|
| ATCC 10031 + OMVs-MEM(+) | 500 | (9.79 ± 3.52) × 10−9 |
| NTUH-K2044 + OMVs-MEM(+) | 500 | (7.83 ± 3.48) × 10−9 |
| ATCC 700603 + OMVs-MEM(+) | 200 | (8.08 ± 1.89) × 10−7 |
| ATCC 700603 + OMVs-MEM(+) | 20 | (1.65 ± 0.25) × 10−7 |
| ATCC 700603 + OMVs-MEM(−) | 20 | (1.42 ± 0.27) × 10−6 |
FIG 3.
HGT via OMVs derived from CRKP. (A) Colony PCR detection of blaNDM-1 from three groups of transformants [ATCC 10031 coincubation with OMVs-MEM(+), ATCC 700603 coincubation with OMVs-MEM(+), and NTUH-K2044 coincubation with OMVs-MEM(+)]. Two colonies were randomly selected from each of the three groups for the detection of blaNDM-1. Genomic DNA of CRKP was used as the positive control, and genomic DNA of ATCC 10031, ATCC 700603, and NTUH-K2044 and sterile water were used as the negative controls (NC). (B) REP-PCR profile of the donor, recipient, and transformant strains used in this study. T, transformant.
Repetitive extragenic palindromic PCR (REP-PCR) was performed on the donors, recipients, and transformants. There was no difference in the REP-PCR amplification profiles between recipient strains and transformants (Fig. 3B). Therefore, contamination by the donor strain was excluded.
Antibiotic sensitivity tests were performed on donors, recipients, and transformants. The CRKP strain showed high β-lactam resistance, including cephalosporin and carbapenem resistance. The transformants showed higher MIC values for β-lactam antibiotics than their respective recipient strains (Table 2). Moreover, there was no difference in the MIC values of ciprofloxacin, indicating that the transformants did not acquire aac(6′)-Ib-cr.
TABLE 2.
Antibiotic susceptibility profiles of donor, recipient and transformant strains
| Antibiotica | MIC (μg/mL) for strain: |
||||||
|---|---|---|---|---|---|---|---|
| CRKP | ATCC 10031c | T-ATCC 10031b | ATCC 700603d | T-ATCC 700603 | NTUH-K2044e | T-NTUH-K2044 | |
| AMP | ≥32 | 16 | ≥32 | ≥32 | ≥32 | ≥32 | ≥32 |
| AMC | ≥32 | ≤2 | ≥32 | 4 | ≥32 | ≤2 | ≥32 |
| TZP | ≥128 | ≤4 | 64 | 8 | 64 | ≤4 | 64 |
| CFZ | ≥64 | ≤4 | ≥64 | ≤4 | ≥64 | ≤4 | ≥64 |
| FOX | ≥64 | ≤4 | ≥64 | ≥64 | ≥64 | ≤4 | ≥64 |
| CRO | ≥64 | ≤1 | ≥64 | ≤1 | ≥64 | ≤1 | ≥64 |
| FEP | ≥64 | ≤1 | 2 | ≤1 | 8 | ≤1 | 2 |
| ATM | ≥64 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| MEM | 16 | ≤0.5 | 4 | ≤0.5 | 4 | ≤0.5 | 4 |
| ETP | ≥8 | ≤0.5 | 4 | ≤0.5 | ≥8 | ≤0.5 | 4 |
| IPM | 4 | ≤1 | 4 | ≤1 | 4 | ≤1 | 4 |
| AMK | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 |
| GEN | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| TOB | 8 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| CIP | 1 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| LVX | 1 | ≤0.25 | ≤0.25 | 1 | 1 | ≤0.25 | ≤0.25 |
| TGC | 2 | ≤0.5 | ≤0.5 | ≥8 | ≥8 | ≤0.5 | ≤0.5 |
| NIT | 64 | ≤16 | ≤16 | 64 | 64 | 64 | 64 |
| SXT | ≥320 | ≤20 | ≤20 | ≤20 | ≤20 | ≤20 | ≤20 |
AMP, ampicillin; AMC, amoxicillin-clavulanic acid; TZP, piperacillin-tazobactam; CFZ, cefazolin; FOX, cefoxitin; CRO, ceftriaxone; FEP, cefepime; ATM, aztreonam; MEM, meropenem; ETP, ertapenem; IPM, imipenem; AMK, amikacin; GEN, gentamicin; TOB, tobramycin; CIP, ciprofloxacin; LVX, levofloxacin; TGC, tigecycline; NIT, nitrofurantoin; SXT, trimethoprim-sulfamethoxazole.
T, transformant.
ATCC 10031, K. pneumoniae reference strains.
ATCC 700603 produces β-lactamase SHV-18 and is a CLSI quality control strain for antimicrobial susceptibility testing.
NTUH-K2044, isolated from a Taiwanese liver abscess, is widely used as a model strain for hvKP.
Virulence level of NTUH-K2044 transformants.
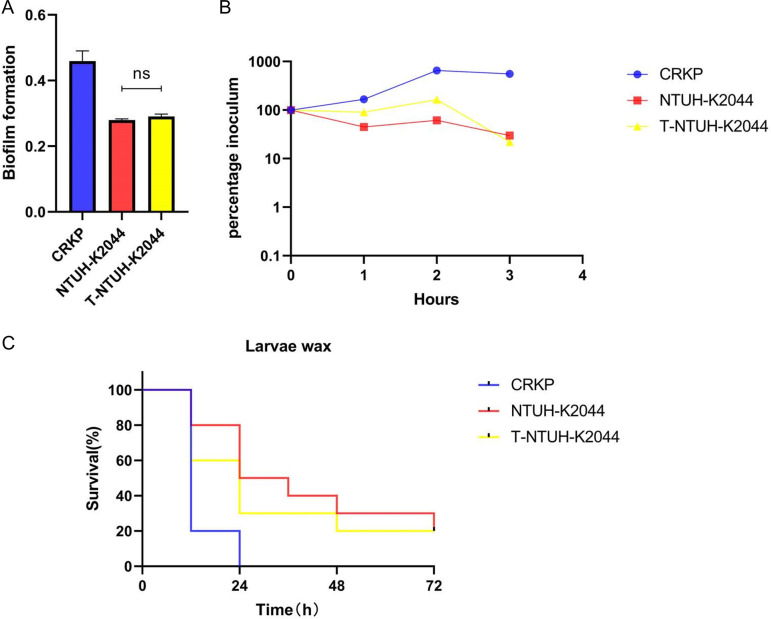
Compared to NTUH-K2044, NTUH-K2044 transformants harboring blaNDM-1 showed no significant difference in biofilm formation (P > 0.05) or serum resistance (Fig. 4A and B), and high virulence was supported by larva infection experiments (Fig. 4C). These data confirmed that the virulence of transformants that acquired drug-resistant genes was not reduced compared to that of NTUH-K2044 recipient bacteria.
FIG 4.
Virulence phenotype of donor, recipient, and transformant strains. (A and B) Biofilm formation (A) and serum resistance (B) of CRKP, NTUH-K2044, and NTUH-K2044 transformants. (C) Virulence level of different strains as depicted in a wax moth larva infection model. ns, P > 0.05. T, transformant.
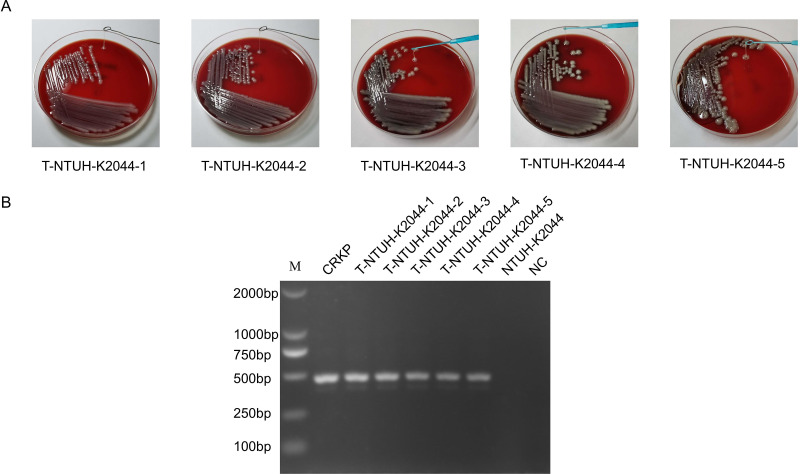
To confirm whether the NTUH-K2044 transformants exhibited a stable hypermucoviscosity phenotype, NTUH-K2044 transformants were passaged for five generations and the hypermucoviscosity phenotype of each of these generations was measured. String tests on blood agar plates showed that blaNDM-1-positive NTUH-K2044 still exhibited a hypermucoviscosity phenotype (Fig. 5A). The blaNDM-1 of colonies from these generations was analyzed using PCR, which showed that the transformants that acquired the resistance genes still maintained blaNDM-1 after several generations (Fig. 5B).
FIG 5.
NTUH-K2044 transformants exhibited a dual phenotype of hypermucoviscosity and carbapenem resistance after several generations. (A) String tests on blood agar plates of five generations of NTUH-K2044 transformants. (B) Colony PCR detection of blaNDM-1 of five generations of NTUH-K2044 transformants. T, transformant.
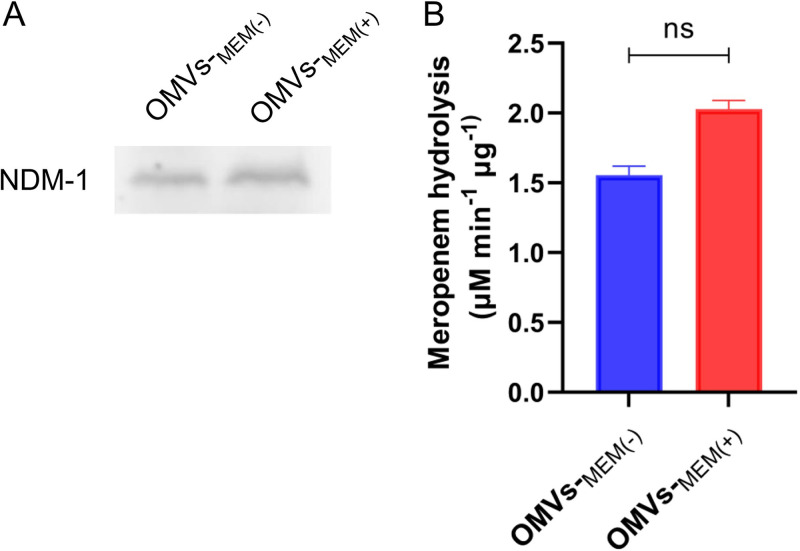
OMVs from CRKP can carry NDM-1 and hydrolyze meropenem.
To verify whether OMVs isolated from CRKP carry metallo-β-lactamases, Western blot analysis was performed using rabbit anti-NDM-1 antibodies. Results showed that OMVs-MEM(−) and OMVs-MEM(+) contained NDM-1 (Fig. 6A). To determine the NDM-1 activity, meropenem was used as the substrate, and the hydrolysis rates of OMVs-MEM(−) and OMVs-MEM(+) were 1.556 ± 0.052 μM min−1 μg−1 and 2.028 ± 0.052 μM min−1 μg−1, respectively (Fig. 6B).
FIG 6.
(A) Immunodetection of NDM-1 in OMVs produced in LB broth without or with 8 μg/mL meropenem [OMVs-MEM(−) and OMVs-MEM(+), respectively] using Western blotting. (B) Meropenem hydrolysis by OMVs-MEM(−) and OMVs-MEM(+) (n = 3). ns, P > 0.05.
DISCUSSION
The World Health Organization (WHO) antimicrobial resistance surveillance report addressed the severity of CRKP, which has a prevalence of >50% in some regions (18). In China, NDM carbapenemase is the second most prevalent carbapenemase after KPC-2 (19). The exchange of genes is common between bacterial species, leading to the rapid spread of resistance. Some features of hvKP make it difficult to acquire genetic material from the environment, including genetic restrictions and the constraints of the significantly thickened capsule. Nevertheless, carbapenem resistance genes have gradually spread to hvKP, leading to the emergence of CR-hvKP (5–7); this represents a significant challenge for antimicrobial therapy. An improved understanding of the mechanisms underlying the spread of resistance genes is needed to limit this serious threat to global health.
In addition to the three classical mechanisms of HGT (conjunction, transformation, and transduction), OMVs are a recently proposed mechanism that might allow HGT. Studies have reported that OMVs carrying resistance and virulence genes have the potential to be delivered to other bacteria (16, 17, 20, 21, 22). Moreover, a recent study has demonstrated that K. pneumoniae can transfer virulence genes to carbapenem-resistant bacteria via OMVs (23). However, OMVs derived from K. pneumoniae that transfer carbapenem resistance genes to hypervirulent K. pneumoniae have been insufficiently investigated.
In this study, OMVs were isolated from CRKP without or with 8-μg/mL meropenem treatment [OMVs-MEM(−) and OMVs-MEM(+), respectively]. The TEM and DLS results showed that both OMVs-MEM(−) and OMVs-MEM(+) were spherical in structure and had no substantial differences in morphology. However, the mean particle size of OMVs-MEM(+) (161.77 nm) was larger than that of OMVs-MEM(−) (147.30 nm), and the protein as well as DNA content of OMVs-MEM(+) was also higher. These differences can be attributed to the antibiotic selection pressure. According to a previous study (24), imipenem enhances the release of OMVs from multidrug-resistant K. pneumoniae, indicating that the generation of OMVs depends on environmental factors. In this study, the protein profiles showed that OMVs-MEM(+) had a protein band at approximately 70 kDa which was substantially elevated compared to that of OMVs-MEM(−) (Fig. 2C). This is in line with the previously presented results (24). The composition and function of the 70-kDa protein identified in the present study require further investigation.
Our experimental results showed that OMVs derived from CRKP could mediate the transfer of blaNDM-1 to ckp, generating CRKP and therefore increasing the carbapenem MIC of the transformants. This demonstrated that, in addition to blaKPC-2 (23), blaNDM-1 could also be transferred between K. pneumoniae cells via OMVs. Another important and novel finding from our study is the transfer of blaNDM-1 to hvKP, indicating the potential for the emergence of CR-hvKP via OMV-mediated HGT. The frequency of OMV-mediated blaNDM-1 transfer to hvKP was slightly lower than that to ckp, suggesting that the gene transfer frequency may be related to the characteristics of the recipient strains. Further exploration is needed regarding the factors affecting the frequency of OMV-mediated transfer of drug-resistant plasmids, like the nature of the capsule polysaccharide. Although more OMVs were formed after meropenem treatment, the frequency of blaNDM-1 gene transfer was not increased compared to that in the untreated group, indicating that the OMVs produced by CRKP following meropenem treatment may have additional, unidentified roles. In addition, despite the inclusion of multiple drug resistance genes in the OMVs, we found that only blaNDM-1 was transferred to the recipient strains, suggesting a selective mechanism that remains to be further investigated.
A previous study (25) reported that the CR-hvKP strain formed by conjugative transfer of the KPC plasmid to NTUH-K2044 presented as nonmucoid colonies in the third generation. Compared with the transconjugant mucoid strain, the nonmucoid strain showed a much higher level of resistance to carbapenems but exhibited significantly decreased virulence levels. However, the CR-hvKP strain formed in our study retained the hypermucoviscosity phenotype throughout the five passages (Fig. 5A). The NTUH-K2044 transformants showed no decrease in biofilm formation and serum resistance, and larva infection experiments showed that these strains retained high virulence. Therefore, these results demonstrated that the CR-hvKP strain formed by OMV-mediated transfer of blaNDM-1 had the potential for a dual phenotype of carbapenem resistance and hypervirulence without significant burdens.
Previous studies have revealed that OMVs secreted from antibiotic-resistant bacteria act as efficient and protective β-lactamase and NDM-1 transporters and can contribute to antibiotic resistance in a nongenetic manner in the bacterial community (26, 27). Our results showed that OMVs derived from CRKP can carry NDM-1 and hydrolyze meropenem. The simultaneous secretion of proteins and genetic elements suggests that OMVs contribute to the spread of antimicrobial resistance, not only by the inactivation of antibiotics through enzyme transport but also by transferring genetic elements to antibiotic-sensitive bacteria. This indicates that new therapeutic options that interfere with vesicle secretion should be explored.
The main limitation of this study is that the mechanism of DNA wrapping into OMVs and the mechanism by which vesicles enter recipient bacterial strains remain to be elucidated. Our future studies will aim to study the selective mechanism underlying transfer of blaNDM-1 and shed light on whether CR-hvKP has the potential to continuously disseminate drug resistance genes.
In summary, our study demonstrated for the first time that OMVs derived from CRKP could carry blaNDM-1 and deliver resistance genes to other K. pneumoniae strains, even hvKP. The transfer of carbapenem resistance genes into hypervirulent strains may promote the emergence and dissemination of CR-hvKP. This study elucidates a new mechanism of HGT which increases the risk of generating CR-hvKP, a concerning development in the fight against antimicrobial resistance.
MATERIALS AND METHODS
Bacterial strains.
CRKP carrying the blaNDM-1 gene was used for the isolation and purification of OMVs. This clinical strain was isolated from the blood of a pediatric patient in 2018 from a tertiary hospital. This strain is reported to harbor blaNDM-1, blaCTX-M-15, blaSHV-12, blaTEM-1C, blaDHA-1, and aac(6′)-Ib-cr resistance determinants based on the whole-genome sequence (28). The CRKP genome contains a chromosome of 5,344,963 bp and four plasmids, pCRKP-1 to -4, with sizes of 262,673 bp, 162,879 bp, 124,316 bp, and 3,681 bp, respectively. Plasmid pCRKP-2 belongs to the IncFIBpKPHS1/IncX3 replicon and harbors β-lactam resistance genes blaNDM-1 and blaSHV-12; blaSHV-12 is not in the neighboring region with blaNDM-1. Plasmid pCRKP-1 contains blaDHA-1, while pCRKP-3 contains blaCTX-M-15, blaTEM-1C, and aac(6′)-Ib-cr. The genome sequence of CRKP was deposited in GenBank (accession number SAMN33420447).
K. pneumoniae ATCC 10031, ESBL-producing K. pneumoniae ATCC 700603, and hvKP NTUH-K2044 were used as recipient strains for transformation experiments.
OMV purification.
CRKP was incubated in Luria-Bertani (LB) medium (Sangon Biotech, China) without antibiotics or with 8 μg/mL meropenem at 37°C and 200 rpm for 12 h and then used for the preparations of OMVs-MEM(−) and OMVs-MEM(+), respectively. Cell cultures were centrifuged at 4°C and 3,000 × g for 15 min, and supernatants were filtered through a 0.22-μm membrane (Merck Millipore, Germany) to remove small cell debris. The filtrates were concentrated using a 100-kDa 50-mL ultrafiltration tube (Millipore) and then subjected to ultracentrifugation at 4°C and 100,000 × g for 3 h using a 70 Ti rotor (Beckman, USA). The pellets were washed with sterile phosphate-buffered saline (PBS) (pH 7.4). Ultracentrifugation was repeated to obtain OMVs. The pellet containing OMVs was resuspended in 200 μL of PBS, and OMVs were cultured on blood agar plates to ensure sterility. The OMVs were stored at −80°C until used.
Quantification of OMV proteins and DNA in OMVs.
OMV protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Beyotime Biotechnology, China), according to the manufacturer’s protocol. Intravesicular DNA was quantified using a previously described method (20) with a few modifications. Briefly, 10 μg of OMVs was treated with 1 U DNase I (Thermo Scientific, USA) and 100 μg/mL proteinase K (Tiangen Biotech, China). DNase I- and proteinase K-treated vesicles were subsequently lysed with 0.125% Triton X-100 (Solarbio, China) solution for 30 min at 37°C, before the DNA was recovered using a DNA purification kit (Vazyme, China), according to the manufacturer’s protocol. The DNA concentration and purity were determined using a NanoDrop spectrophotometer (Thermo Scientific).
TEM.
Purified OMVs were visualized using a transmission electron microscope (TEM; TEM-1400Plus; Japan) with negative staining. OMVs (30 μL) were absorbed onto a carbon-coated Formvar film and negatively stained with 2.0% (wt/vol) uranyl acetate at room temperature for 5 min. Samples were observed under a TEM operating at 120 kV.
DLS.
The particle size and polydispersity index of the OMVs were determined using a Zeta-sizer nano-ZS (Malvern Instruments, UK). OMV samples were diluted to a concentration of 20 μg/mL with sterile PBS buffer, and 1 mL of these OMV aliquots was pipetted into a sterile cuvette. All measurements were performed at 25°C, and experiments were performed in triplicate.
PCR.
The presence of resistance genes [blaNDM-1, blaCTX-M-15, blaSHV-12, blaTEM-1C, blaDHA-1, and aac(6′)-Ib-cr] was detected using PCR. The genomic DNA of K. pneumoniae was extracted using the boiling method. The PCR products were visualized on a 1% agarose gel. Primers used in this study are listed in Table 3.
TABLE 3.
Oligonucleotides used in this study
| Primer | Sequence (5′–3′) | Length (bp) | Use |
|---|---|---|---|
| NDM-1 Forw | GGGCAGTCGCTTCCAACGGT | 476 | PCR |
| NDM-1 Rev | GTAGTGCTCAGTGTCGGCAT | ||
| CTX-M-15 Forw | TTGTTAGGAAGTGTGCCGCT | 819 | PCR |
| CTX-M-15 Rev | TTACAAACCGTCGGTGACGA | ||
| SHV-12 Forw | TCTCCCTGTTAGCCACCCT | 793 | PCR |
| SHV-12 Rev | ATTTGCTGATTTCGCTCGGC | ||
| TEM Forw | GTGCGCGGAACCCCTATT | 919 | PCR |
| TEM Rev | TTACCAATGCTTAATCAGTGAGGC | ||
| DHA Forw | AACTTTCACAGGTGTGCTGGGT | 405 | PCR |
| DHA Rev | CCGTACGCATACTGGCTTTGC | ||
| aac(6′)-Ib-cr Forw | ATATGCGGATCCAATGAGCAACGCAAAAACAAAGTTAG | 544 | PCR |
| aac(6′)-Ib-cr Rev | ATAGCGAATTCTTAGGCATCACTGCGTGTTCGCTC | ||
| REP Forw | IIIGCGCCGICATCAGGC | –a | REP-PCR |
| REP Rev | ACGTCTTATCAGGCCTAC |
no specific length.
OMV-mediated blaNDM-1 transfer.
OMV-mediated blaNDM-1 transfer experiments were performed as previously described (21), with slight modifications. ckp ATCC 10031, ckp ATCC 700603, and hvKP NTUH-K2044 (recipient strains) were grown in LB broth at 37°C and 200 rpm for 4 to 5 h to an optical density at 600 nm (OD600) of 0.4. Cells were centrifuged and resuspended in cold LB broth to a final concentration of 107 CFU/mL. These suspensions (100 μL) were mixed with OMV-MEM(−) or OMV-MEM(+) and incubated statically at 37°C for 4 h, followed by another 4 h of shaking (180 rpm). LB broth was added to a final volume of 10 mL, and the incubation was continued overnight at 37°C with shaking (180 rpm). Bacterial cultures were centrifuged and resuspended in 1 mL of fresh LB broth. A 100-μL aliquot of the suspension was incubated overnight on MAC agar (for total bacterial counts) or MAC agar supplemented with 1 μg/mL meropenem (for transformant selection). The frequency of OMV-mediated blaNDM-1 transfer was calculated as the number of transformants (CFU per milliliter) divided by the total bacterial count (CFU per milliliter). Two randomly selected transformants from each recipient were confirmed to harbor blaNDM-1 by PCR.
To demonstrate that DNA transfer was mediated by OMVs, two sets of experiments were performed: (i) plasmid DNA isolated from CRKP and (ii) OMVs lysed using 0.125% Triton X-100. All experiments were performed in duplicate.
REP-PCR.
Repetitive extragenic palindromic PCR (REP-PCR) was performed as previously described (29). The primers used to perform REP-PCR are listed in Table 3. Two strains were considered to belong to the same genotype if they had the same pattern of bands, and a maximum difference of two band patterns was considered acceptable.
Antibiotic susceptibility testing.
Antibiotic susceptibility testing of the donor, recipient, and transformant strains was performed using broth microdilution. The MICs of ampicillin, amoxicillin-clavulanic acid, piperacillin-tazobactam, cefazolin, cefoxitin, ceftriaxone, cefepime, aztreonam, meropenem, ertapenem, imipenem, amikacin, gentamicin, tobramycin, ciprofloxacin, levofloxacin, tigecycline, nitrofurantoin, and trimethoprim-sulfamethoxazole were determined according to the Clinical and Laboratory Standards Institute (30).
Biofilm formation assay.
Biofilm formation assays were implemented as previously described (31), with slight modifications. In brief, 200 μL of bacterial culture (1.5 × 106 CFU/mL) was added to 96-well flat-bottomed plates and incubated for 24 h at 37°C. Cultures were subsequently removed and washed three times with PBS. The wells were dried, stained with 1% crystal violet for 15 min, and then washed three times with distilled water. The absorbance was measured at 570 nm after solubilization with 95% ethanol. Each assay was performed in duplicate and independently repeated three times.
Serum resistance assay.
The serum resistance experiments were performed as previously described (31). Bacteria grown to the mid-log phase were mixed with normal human serum in a 1:3 ratio and then incubated at 37°C for 3 h. At 1-h intervals following incubation, serially diluted bacteria were plated on Mueller-Hinton agar. Serum resistance was determined by plotting the survival percentage of each strain against the incubation time. All experiments were performed in triplicate.
Larva infection models.
Galleria mellonella larvae weighing 200 to 400 mg, purchased from Tianjin Huiyude Biotechnology Company, were used to determine the virulence levels of donor, recipient, and transformant strains. The concentration of the strain was adjusted to 1 × 108 CFU/mL. Each group of 10 G. mellonella larvae was injected with 10 μL of bacterial culture (106 CFU) and incubated at 37°C for 72 h, and the survival rates of the larvae were observed every 12 h. Animal experiments were performed in duplicate.
String test.
The NTUH-K2044 transformant strain was incubated on a blood agar plate at 37°C overnight. A positive string test was performed by stretching a viscous string with an inoculation loop; viscous strings with a length of >5 mm indicate a hypermucoviscosity phenotype.
SDS-PAGE, NDM-1 detection, and β-lactamase activity.
Sixteen-microgram OMV samples were mixed with 5× loading buffer, denatured by heating at 100°C for 10 min, and loaded onto 10% SDS-PAGE gels. The gels were stained with Coomassie brilliant blue (Sangon Biotech, China), or the separated proteins were transferred onto the polyvinylidene difluoride (PVDF) membrane. Western blotting using the polyclonal anti-NDM-1 antibody at a 1:1,000 dilution (Novus Biologicals) and goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated secondary antibody was performed as previously described (32, 33), with slight modifications. Chemiluminescent signals were acquired using an Image Quant 800 (GE Healthcare).
The metallo-β-lactamase activity was determined by monitoring the absorbance using 256 μg/mL meropenem as the substrate in a reaction buffer containing 10 mM HEPES and 200 mM NaCl (pH 7.4). One microgram of OMVs was added into reaction mixtures, for a total volume of 100 μL, and the absorbance at 300 nm was measured immediately in kinetic mode for 10 min. A standard curve was generated using 0 to 256 μg/mL meropenem. The NDM-1 activity of the sample was expressed as the number of micromoles of meropenem hydrolyzed per minute per microgram of protein.
Ethics statement.
Isolates were collected from routine diagnoses, and only pure cultures of bacterial isolates were investigated. No patient-related data were analyzed. According to the regulations of Xiangya Hospital of Central South University, such research does not require ethical approval.
Data availability.
The sequence of the whole genome of the NTUH-K2044 transformants was deposited in GenBank under accession number SAMN33420516. The genome sequence of CRKP was deposited in GenBank under accession number SAMN33420447.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2018YFC2000203).
We thank Taiwan University for the donation of NTUH-K2044.
There are no conflicts of interest to declare.
Contributor Information
Xianghui Liang, Email: Liangxh901@csu.edu.cn.
Wenen Liu, Email: wenenliu@163.com.
REFERENCES
- 1.Wang M, Earley M, Chen L, Hanson BM, Yu Y, Liu Z, Salcedo S, Cober E, Li L, Kanj SS, Gao H, Munita JM, Ordoñez K, Weston G, Satlin MJ, Valderrama-Beltrán SL, Marimuthu K, Stryjewski ME, Komarow L, Luterbach C, Marshall SH, Rudin SD, Manca C, Paterson DL, Reyes J, Villegas MV, Evans S, Hill C, Arias R, Baum K, Fries BC, Doi Y, Patel R, Kreiswirth BN, Bonomo RA, Chambers HF, Fowler VG, Jr, Arias CA, van Duin D, Multi-Drug Resistant Organism Network Investigators . 2022. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis 22:401–412. doi: 10.1016/S1473-3099(21)00399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russo TA, Marr CM. 2019. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 32:e00001-19. doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X, Dong N, Chan EW, Zhang R, Chen S. 2021. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol 29:65–83. doi: 10.1016/j.tim.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Yan R, Lu Y, Zhu Y, Lan P, Jiang S, Lu J, Shen P, Yu Y, Zhou J, Jiang Y. 2021. A sequence type 23 hypervirulent Klebsiella pneumoniae strain presenting carbapenem resistance by acquiring an IncP1 bla KPC-2 plasmid. Front Cell Infect Microbiol 11:641830. doi: 10.3389/fcimb.2021.641830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. 2016. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother 60:709–711. doi: 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cejas D, Fernandez Canigia L, Rincon Cruz G, Elena AX, Maldonado I, Gutkind GO, Radice MA. 2014. First isolate of KPC-2-producing Klebsiella pneumoniae sequence type 23 from the Americas. J Clin Microbiol 52:3483–3485. doi: 10.1128/JCM.00726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Long D, Xiang TX, Du FL, Wei DD, Wan LG, Deng Q, Cao XW, Zhang W. 2019. Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J Antimicrob Chemother 74:1233–1240. doi: 10.1093/jac/dkz023. [DOI] [PubMed] [Google Scholar]
- 8.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 9.Shen D, Ma G, Li C, Jia X, Qin C, Yang T, Wang L, Jiang X, Ding N, Zhang X, Yue L, Yin Z, Zeng L, Zhao Y, Zhou D, Chen F. 2019. Emergence of a multidrug-resistant hypervirulent Klebsiella pneumoniae sequence type 23 strain with a rare blaCTX-M-24-harboring virulence plasmid. Antimicrob Agents Chemother 63:e02273-18. doi: 10.1128/AAC.02273-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua Y, Wang J, Huang M, Huang Y, Zhang R, Bu F, Yang B, Chen J, Lin X, Hu X, Zheng L, Wang Q. 2022. Outer membrane vesicles-transmitted virulence genes mediate the emergence of new antimicrobial-resistant hypervirulent Klebsiella pneumoniae. Emerg Microbes Infect 11:1281–1292. doi: 10.1080/22221751.2022.2065935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall RJ, Whelan FJ, McInerney JO, Ou Y, Domingo-Sananes MR. 2020. Horizontal gene transfer as a source of conflict and cooperation in prokaryotes. Front Microbiol 11:1569. doi: 10.3389/fmicb.2020.01569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ely B. 2020. Recombination and gene loss occur simultaneously during bacterial horizontal gene transfer. PLoS One 15:e0227987. doi: 10.1371/journal.pone.0227987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redondo-Salvo S, Fernandez-Lopez R, Ruiz R, Vielva L, de Toro M, Rocha EPC, Garcillan-Barcia MP, de la Cruz F. 2020. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat Commun 11:3602. doi: 10.1038/s41467-020-17278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolling GL, Matthews KR. 1999. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol 65:1843–1848. doi: 10.1128/AEM.65.5.1843-1848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dell’Annunziata F, Dell’Aversana C, Doti N, Donadio G, Dal Piaz F, Izzo V, De Filippis A, Galdiero M, Altucci L, Boccia G, Folliero V, Franci G. 2021. Outer membrane vesicles derived from Klebsiella pneumoniae are a driving force for horizontal gene transfer. Int J Mol Sci 22:8732. doi: 10.3390/ijms22168732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 19.Han R, Shi Q, Wu S, Yin D, Peng M, Dong D, Zheng Y, Guo Y, Zhang R, Hu F, China Antimicrobial Surveillance Network (CHINET) Study Group . 2020. Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol 10:314. doi: 10.3389/fcimb.2020.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatterjee S, Mondal A, Mitra S, Basu S. 2017. Acinetobacter baumannii transfers the blaNDM-1 gene via outer membrane vesicles. J Antimicrob Chemother 72:2201–2207. doi: 10.1093/jac/dkx131. [DOI] [PubMed] [Google Scholar]
- 21.Rumbo C, Fernandez-Moreira E, Merino M, Poza M, Antonio Mendez J, Soares NC, Mosquera A, Chaves F, Bou G. 2011. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother 55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bielaszewska M, Daniel O, Karch H, Mellmann A. 2020. Dissemination of the blaCTX-M-15 gene among Enterobacteriaceae via outer membrane vesicles. J Antimicrob Chemother 75:2442–2451. doi: 10.1093/jac/dkaa214. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Wen Z, Jiang M, Xia F, Wang M, Zhuge X, Dai J. 2022. Dissemination of virulence and resistance genes among Klebsiella pneumoniae via outer membrane vesicle: an important plasmid transfer mechanism to promote the emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Transbound Emerg Dis 69:e2661–e2676. doi: 10.1111/tbed.14615. [DOI] [PubMed] [Google Scholar]
- 24.Ye C, Li W, Yang Y, Liu Q, Li S, Zheng P, Zheng X, Zhang Y, He J, Chen Y, Hua L, Yang Z, Li D, Ren Z, Yang Y, Qi J, Huang W, Ma Y. 2021. Inappropriate use of antibiotics exacerbates inflammation through OMV-induced pyroptosis in MDR Klebsiella pneumoniae infection. Cell Rep 36:109750. doi: 10.1016/j.celrep.2021.109750. [DOI] [PubMed] [Google Scholar]
- 25.Tian D, Liu X, Chen W, Zhou Y, Hu D, Wang W, Wu J, Mu Q, Jiang X. 2022. Prevalence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae under divergent evolutionary patterns. Emerg Microbes Infect 11:1936–1949. doi: 10.1080/22221751.2022.2103454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SW, Park SB, Im SP, Lee JS, Jung JW, Gong TW, Lazarte JMS, Kim J, Seo JS, Kim JH, Song JW, Jung HS, Kim GJ, Lee YJ, Lim SK, Jung TS. 2018. Outer membrane vesicles from beta-lactam-resistant Escherichia coli enable the survival of beta-lactam-susceptible E. coli in the presence of beta-lactam antibiotics. Sci Rep 8:5402. doi: 10.1038/s41598-018-23656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez MMB, Bonomo RA, Vila AJ, Maffia PC, Gonzalez LJ. 2021. On the offensive: the role of outer membrane vesicles in the successful dissemination of New Delhi metallo-beta-lactamase (NDM-1). mBio 12:e0183621. doi: 10.1128/mBio.01836-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pei N, Li Y, Liu C, Jian Z, Liang T, Zhong Y, Sun W, He J, Cheng X, Li H, Lei X, Liu X, Deng Z, Liu Q, Chen X, Yan Q, Kristiansen K, Li J, Liu W. 2022. Large-scale genomic epidemiology of Klebsiella pneumoniae identified clone divergence with hypervirulent plus antimicrobial-resistant characteristics causing within-ward strain transmissions. Microbiol Spectr 10:e0269821. doi: 10.1128/spectrum.02698-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bou G, Cervero G, Dominguez MA, Quereda C, Martinez-Beltran J. 2000. PCR-based DNA fingerprinting (REP-PCR, AP-PCR) and pulsed-field gel electrophoresis characterization of a nosocomial outbreak caused by imipenem- and meropenem-resistant Acinetobacter baumannii. Clin Microbiol Infect 6:635–643. doi: 10.1046/j.1469-0691.2000.00181.x. [DOI] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing. M100-S29. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 31.Choi MJ, Ko KS. 2015. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother 59:6763–6773. doi: 10.1128/AAC.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez LJ, Bahr G, Nakashige TG, Nolan EM, Bonomo RA, Vila AJ. 2016. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-beta-lactamase. Nat Chem Biol 12:516–522. doi: 10.1038/nchembio.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez C, Ayala JA, Bonomo RA, Gonzalez LJ, Vila AJ. 2019. Protein determinants of dissemination and host specificity of metallo-beta-lactamases. Nat Commun 10:3617. doi: 10.1038/s41467-019-11615-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence of the whole genome of the NTUH-K2044 transformants was deposited in GenBank under accession number SAMN33420516. The genome sequence of CRKP was deposited in GenBank under accession number SAMN33420447.