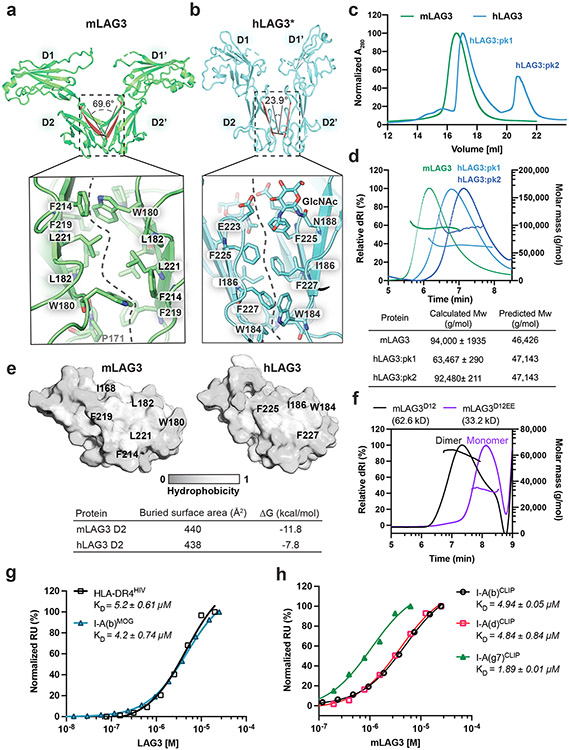
Figure 2. Structural and biochemical characterization of human and murine LAG3 dimers.
a-b, Comparison of mLAG3 and hLAG3 dimer conformations (top). Different D2-D2 packing geometries were observed in mLAG3 (a) and hLAG3 (b). The dihedral angles between D2 beta strands are highlighted in red. The lower panels depict residue networks at the dimer interfaces of mLAG3 (a) and hLAG3 (b). c, SEC chromatograms from hLAG3 and mLAG3 purification. d, SEC-MALS analyses of mLAG3, hLAG3:pk1, and hLAG3:pk2. Calculated and predicted molecular weights are listed in the table. e, Analyses of surface hydrophobicity for D2 domains of mLAG3 (left) and hLAG3 (right); lighter coloring indicates increased surface hydrophobicity. f, SEC-MALS analyses indicate that mLAG3D12 is a homodimer in solution while the mLAG3D12EE mutant is predominantly monomeric. g, SPR was used to determine the binding affinities of hLAG3* and mLAG3 for HLA-DR4HIV and I-A(b)MOG, respectively. h, SPR analyses of the binding affinities of mLAG3 binding to MHCII allomorphs I-A(b), I-A(d) and I-A(g7) loaded with CLIP peptide. The KD and standard deviation were determined by averaging the values from two independent experiments.