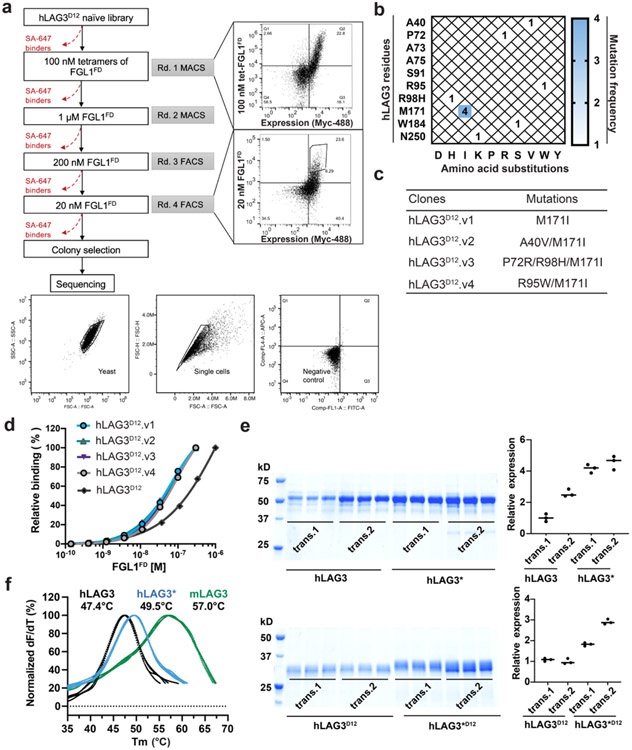
Extended Data Figure 1. Yeast display selections and protein engineering of hLAG3.
a, Yeast display selection strategy used to isolate high-affinity FGL1FD binders from the hLAG3D12 mutant library. Flow cytometry dot plots depict the first round (upper panel) and final round of selection (lower panel) stained with 100 nM FGL1FD tetramers and 20 nM FGL1FD monomers, respectively. The gating strategies for yeast and single cells were shown in the bottom. b, Mutation-frequency map generated from sequencing of the five clones isolated from the final round of yeast selection. The recurring mutation, M171I, is highlighted in blue. c, Table of clones containing the M171I mutation. d, Dose-response titrations of yeast expressing hLAG3D12 or LAG3D12 variants with biotinylated FGL1FD. e, SDS-PAGE analysis of LAG3 protein expression and secretion. Two replicated transfections were performed for hLAG3, hLAG3*, hLAG3D12, hLAG3*D12 (hLAG3D12 with M171I mutation). High-Five cells were inoculated with consistent titer of virus. After 48 hours, proteins were retrieved using Ni-NTA. Quantifications of the relative intensity of the bands (right panel) were based on triplicates inoculation.