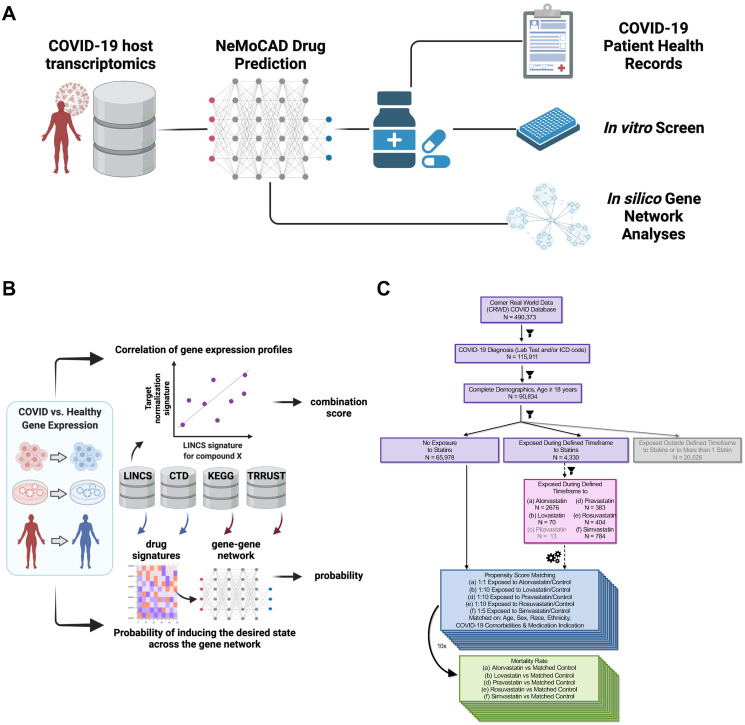
Fig 1. Combination approach for pathway-agnostic identification of compounds for drug repositioning.
(a) Overview of the combination approach to drug repositioning. (b) The NeMoCAD gene network analysis tool is a drug repurposing algorithm that uses Bayesian statistical network analysis combined with data from publicly available datasets (e.g., LINCS, KEGG, CTD, TRRUST) for reference transcriptional signatures and to define regulatory network architecture. The algorithm identifies transcriptome-wide differential expression profiles between two biological states (e.g., healthy vs. diseased) in experimental or published transcriptomic datasets and defines the target normalization signature, i.e., the subset of genes that would need to reverse their expression to revert one state to the other. The output of NeMoCAD includes correlation and causation predictions for numerous chemical compounds and approved drugs in the LINCS database based on their ability to reverse the differential expression profile of interest. (c) Patient selection and analysis of electronic health records. (Figure panels a & b created with BioRender.com.).