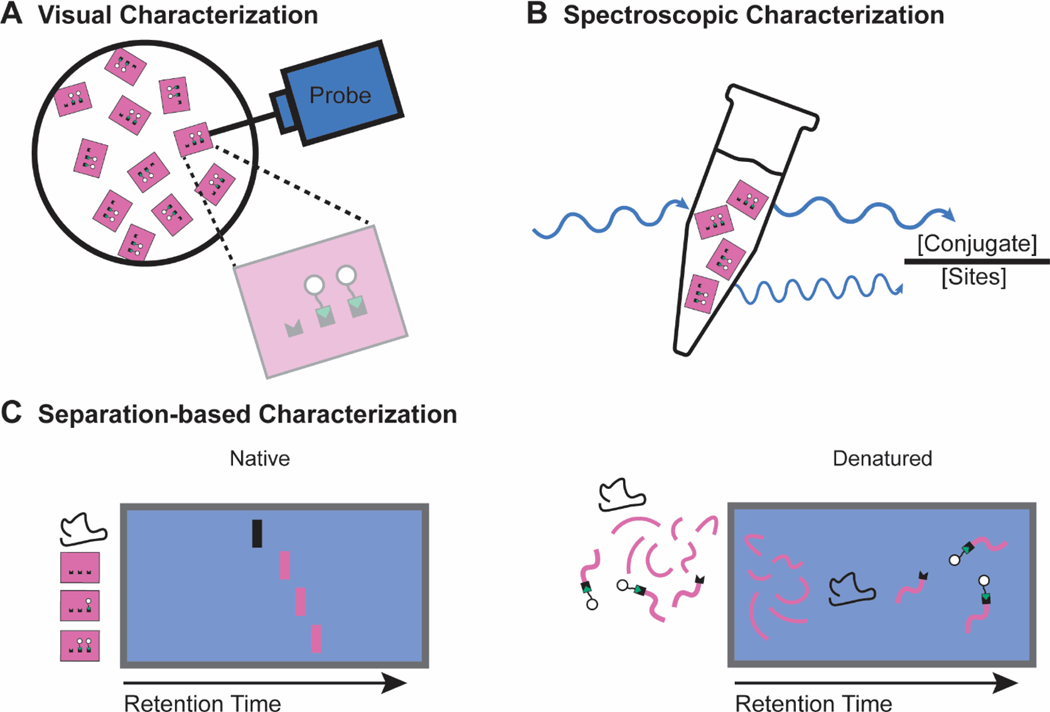
Figure 5. Characterization techniques used to quantify the functionalization of DNA origami nanostructures.
a | Visual characterization techniques rely on a physical probe (light or matter) to create a visual reconstruction of the nanostructure. The images are then used to quantify functionalization efficiency. b | Spectroscopic characterization techniques rely on a spectral fingerprint of the conjugate. UV or fluorescence spectroscopy determines the concentration of conjugate in a sample, which is compared to the concentration of nanostructures for functionalization efficiency quantification. c | Native separation-based characterization utilizes differences in retention time of functionalized nanostructures and unfunctionalized nanostructures to resolve, and in some cases quantify, functionalization efficiency. Denatured separation-based characterization relies on the physical separation and subsequent identification of the nanostructure components. Comparing the amount of reacted and unreacted functionalization sites allows for functionalization efficiency quantification.