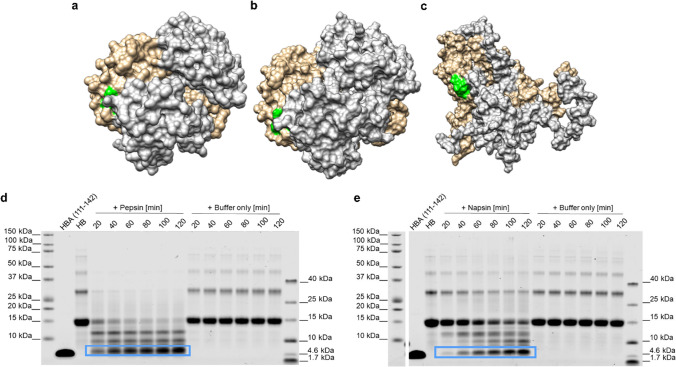
Fig. 3.
Generation of HBA(111–142) by proteolytic digestion of hemoglobin. a Hemoglobin crystal structure (PDB code: 2DN2 [31]). b Representative structure of hemoglobin from the simulations at neutral pH (7.4). c Representative structure of hemoglobin from the simulations at acidic pH (3.6). Two α and two β subunits are shown in brown and gray, respectively. The proposed recognition site (108–113) is displayed in green. Human hemoglobin was incubated with d pepsin (at pH 3.5) or e napsin A (at pH 3.6) at a 1:100 molar ratio, or with the respective digestion buffers without the addition of protease for 20, 40, 60, 80, 100, or 120 min at 37 °C. The reactions were separated by SDS-PAGE and total protein stained by colloidal Coomassie. The peptide of interest is marked with a blue box. Also see Fig. S3