Figure 3. Diversity of OPSCC malignant cells.
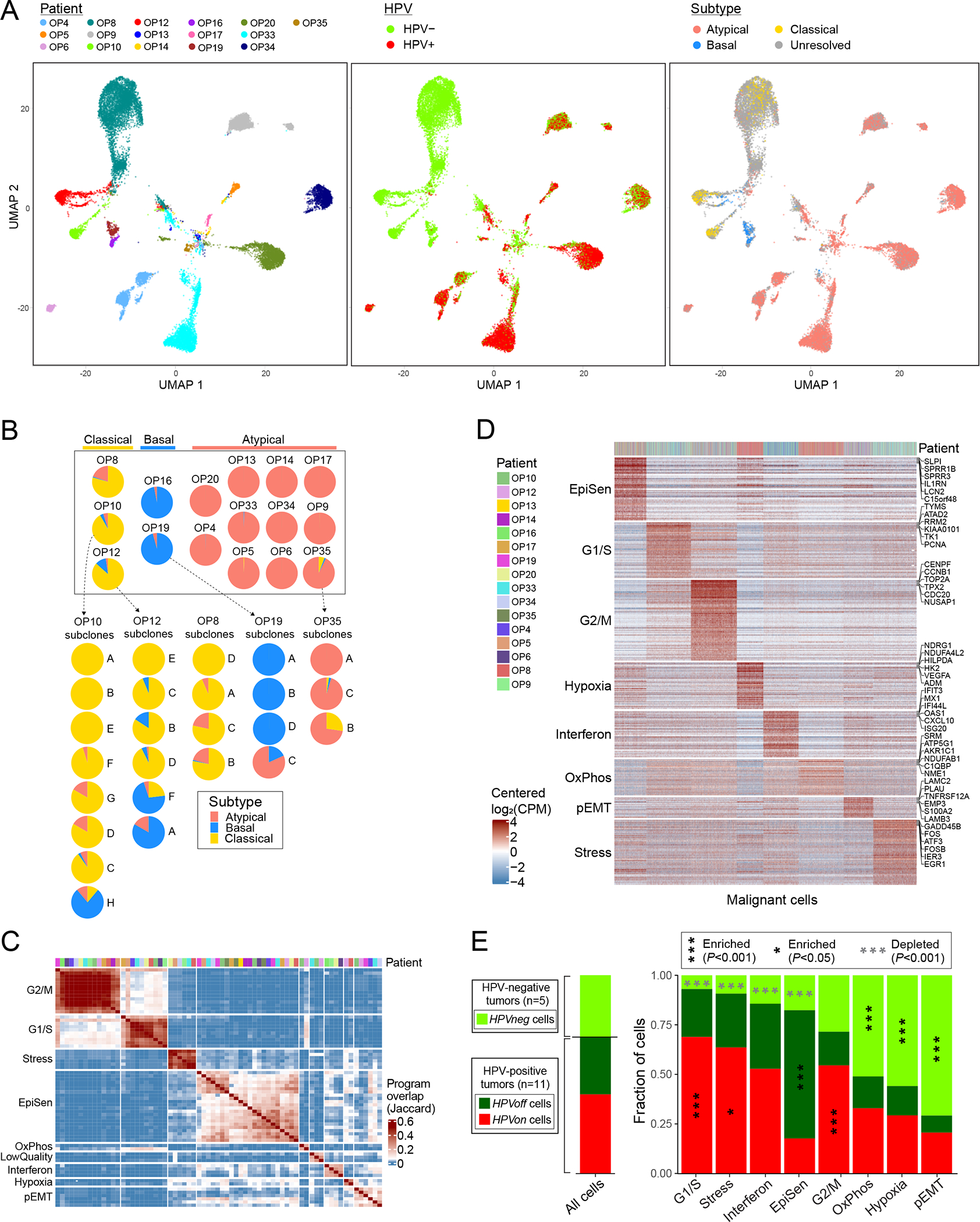
(A) UMAPs of all malignant cells (n=20,323) colored by patient (left panel), HPV expression (middle panel) and TCGA subtype (right panel). Cells with smaller than 1.5 fold-change between the top and the second highest subtype scores were defined as unresolved and marked in grey. (B) Pie charts representing the fraction of cells assigned to each TCGA subtype (excluding unresolved cells), per patient (above) and per subclone for patients with multiple subtypes and multiple subclones (below). (C) Hierarchical clustering of 69 NNMF-derived program signatures from 16 patients (see Methods). Signatures are clustered by Jaccard overlap. Groups of signatures, from which meta-programs are derived, are annotated on the left. Top panel shows the patient origin for each program using the same color map as in (D). (D) Expression of meta-program genes (rows) in all malignant cells (columns). Top panel indicates the patient origin for every cell. (E) For each meta-program, bar-plot shows the fraction of cells, out of those assigned to that meta-program, in three HPV-related classes: cells from HPV-negative tumors (HPVneg, light green) and cells from HPV-positive tumors in which HPV reads are detected (HPVon, red) or undetected (HPVoff, dark green). Asterisks denote enrichment (black and vertical) or depletion (grey and horizontal); asterisks within the HPVneg area denote enrichment/depletion in HPVneg vs. HPV-positive tumors (HPVon and HPVoff), and asterisks within the HPVon or HPVoff area denote enrichment in comparison between those two classes. Significance of enrichment/depletion was calculated using a hypergeometric test, corrected for multiple-testing. Bar-plot at the left shows the same analysis for all malignant cells. When calculating all fractions, 100 cells per patient and subset were randomly sampled 100 times to avoid patients with more cells skewing the results.
