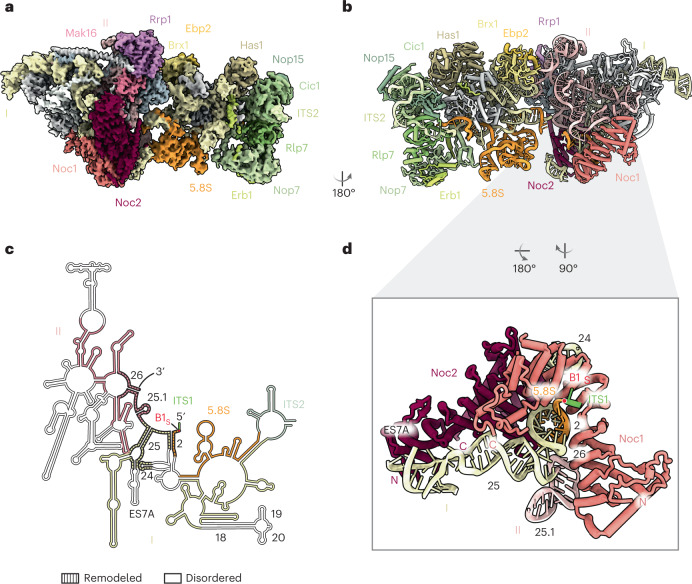
Fig. 1. Cryo-EM structure of the co-transcriptional Noc1–Noc2 RNP.
a, Cryo-EM reconstruction of the Noc1–Noc2 RNP, visualizing domains I and II of the 25S rRNA with ITS2 and the 5.8S rRNA, and associated assembly factors and ribosomal proteins. b, Corresponding atomic model of the Noc1–Noc2 RNP, rotated by 180°. c, rRNA secondary structure diagram of the yeast LSU rRNA visualized in the Noc1–Noc2 RNP cryo-EM reconstruction. rRNA elements present in the structure are color-coded. Disordered segments are colored in white, and remodeled segments are indicated by dashed lines, with relevant helices numbered in black. d, The Noc1–Noc2 dimer clamps around root helix 2 of the pre-rRNA that is formed by base-pairing of the 5.8S and 25S rRNAs, and remodels root helices 25 and 26.