Figure 4: A 2kb deletion within intron 3 of LRRN4 is a strong candidate for causal variant at the chr20 rs77351827 locus.
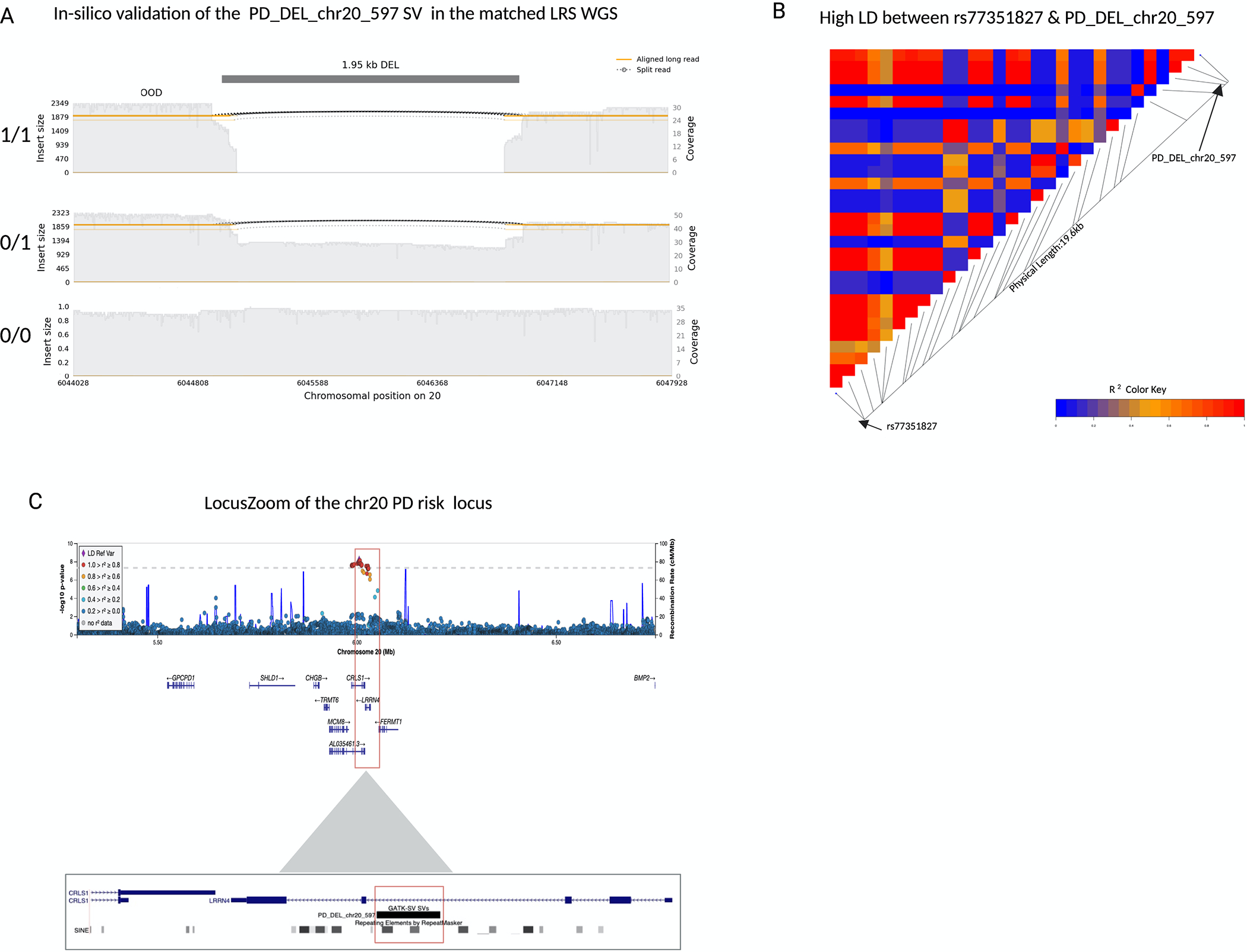
a) A samplot image showing the ~2kb deletion at chr20. Aligned regions are marked in orange and the gap represents the deletion coded in black. The height of the alignment is based on the size of its largest gap. The three sequence alignment tracks follow, each alignment file plotted as a separate track in the image. The coverage for the region is shown with the gray-filled background. The SV genotypes (homozygous deletion, heterozygous deletion, and homozygous reference allele/no deletion) that were predicted by GATK-SV from the short-read sequencing data are annotated on the left of the corresponding tracks. Each genotype was confirmed in-silico by the matched long-read sequencing data. b) A LDheatmap showing pairwise LD measurements measured in R2 between the 2kb PD_DEL_chr20_597 deletion and rs77351827. High R2 values are shown in red and low R2 values in blue. PD_DEL_chr20_597 is in high LD with the lead PD risk SNV of this locus rs77351827(r2=0.89, D’=0.95). c) Locuszoom plot of the association signal at the chr20 rs77351827 PD risk locus from the Nalls 2019 PD SNV meta-analysis. The gene LRRN4 lies directly under the risk signal and the schematic below shows the location of the deletion within intron 3 of the gene.
