Figure 5: Comparison of SVs called with short-read and long-read in eight matched PPMI blood samples -.
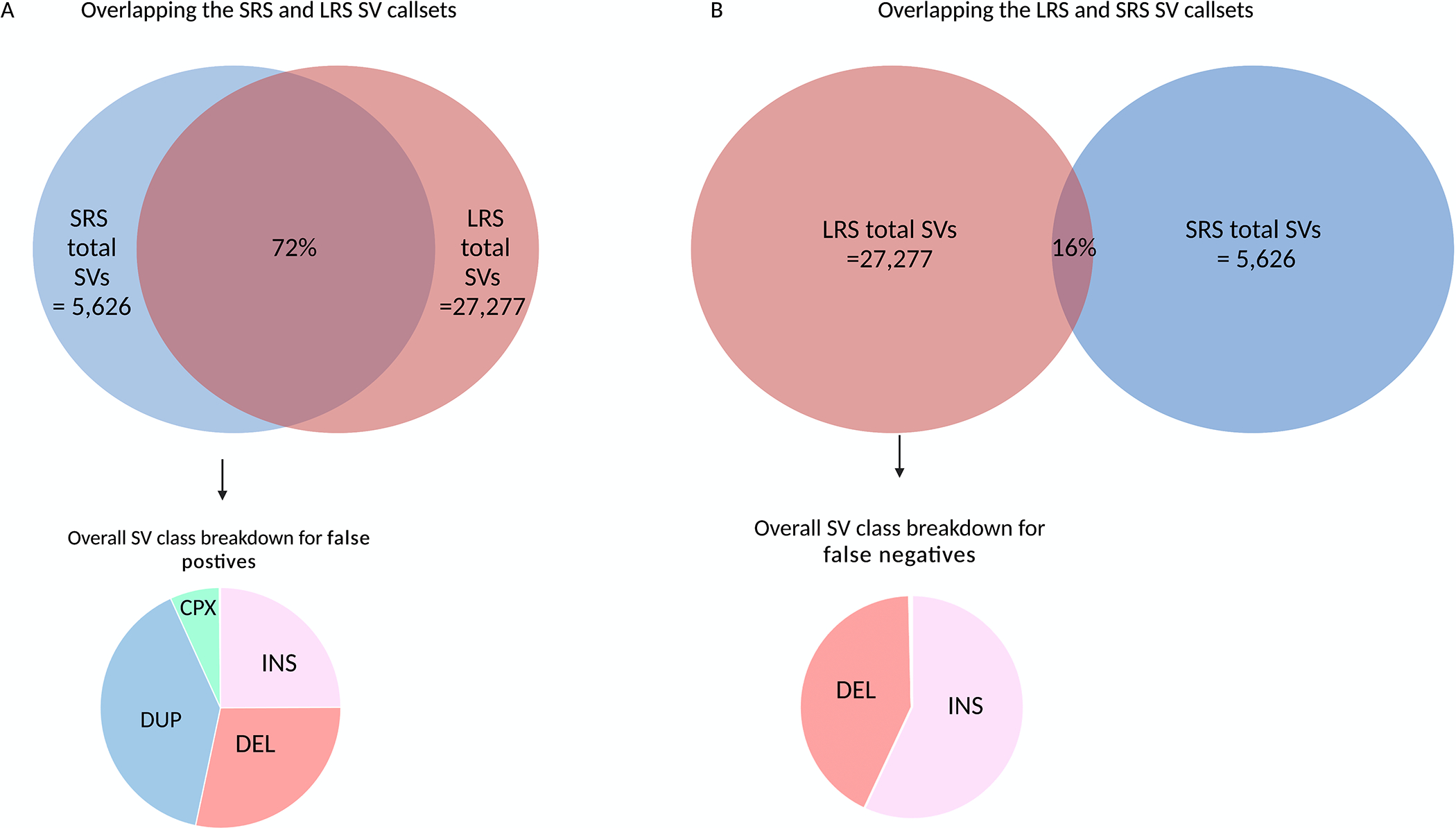
ONT long-read sequencing detects significantly more SV than the short-read sequencing on average per genome. a) On average 5,626 SVs were detected per short-read genome compared to 27,277 with long-read sequencing. Of the 5,626 SV discovered in the short-read sequencing data, 72% of the SV were confirmed in-silico with long-read sequencing. As expected, duplications drove the false positive rate. b) The majority of the SV in the genome cannot be detected with sequencing data alone. Of the 27,277 SVs detected with long-read sequencing, only 14% of the SVs were present in the short-read callset. Most of these false negative calls, i.e SVs that were detected by long-read sequences but not present in the short-read callset were insertions.
