Figure 1. iPAK4 forms intracellular protein fibers.
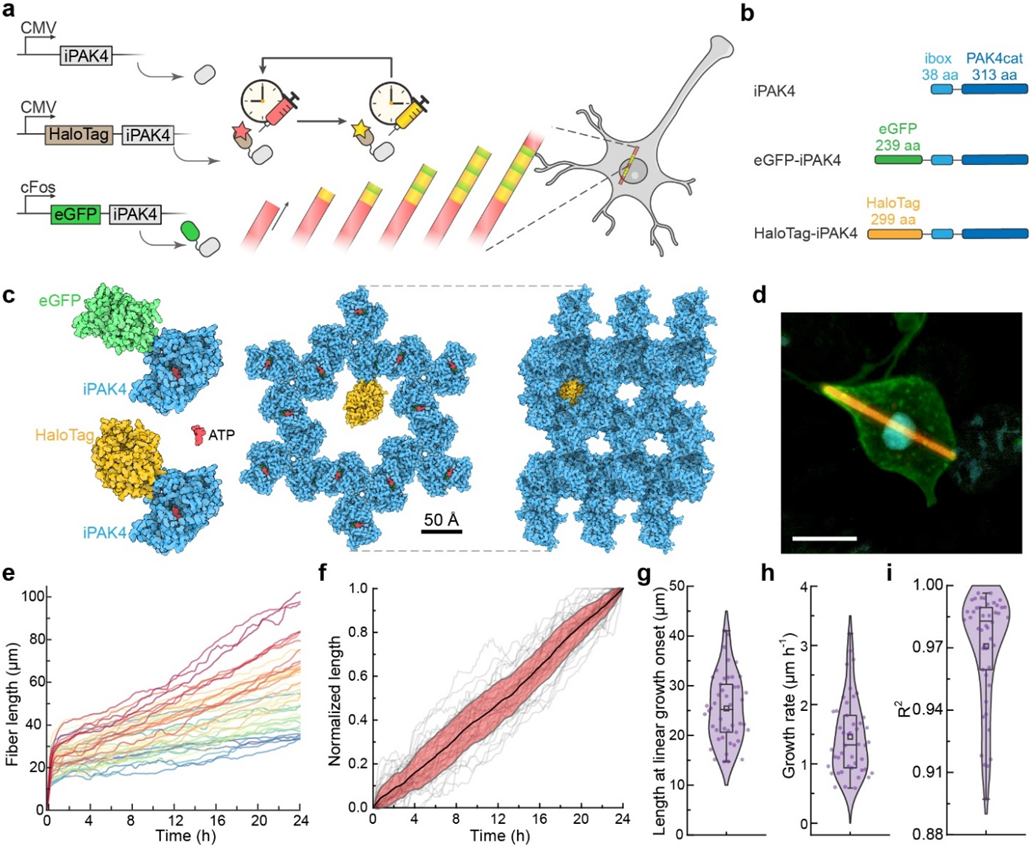
a, Scheme for intracellular recording of cFos activity with fiducial timestamps. iPAK4 forms the fiber scaffold. HaloTag-iPAK4 incorporates HT dyes, permitting labeling of the fiber with fiducial timestamps. Neural activation drives expression of cFos::eGFP-iPAK4, introducing green bands into the fiber. b, Composition of the protein constructs used to label intracellular protein fibers. c, The structures of tagged iPAK4 monomers and the crystal structure with hexagonal pores (from PDB:4XBR) were modeled using Protein Imager software.39 d, Image of a HEK cell expressing CMV::iPAK4 (95%) and CMV::HT-iPAK4 (5%). The fiber was stained with JFX608, the membrane was labeled by expressing GPI-eGFP, and the nucleus was labeled with DAPI. Scale bar 10 μm. e. Growth profiles of iPAK4 fibers in HEK cells (n = 46 fibers). f, Population average (black) and standard deviation (red) of the linear-phase fiber growth (grey lines). Here each fiber’s linear-phase growth profile was mapped to the interval [0, 1]. g-i, Statistics of fiber growth, showing g, fiber length at the transition from initial nucleation to linear growth, h, linear-phase growth rate, and i, R2 of the fit of the linear-phase growth to a straight line. In e-i, fibers were randomly selected from Movie S1. Box bounds: 25th and 75th percentile; whiskers: minimum and maximum; squares: mean; center lines: median. All data points are displayed.
