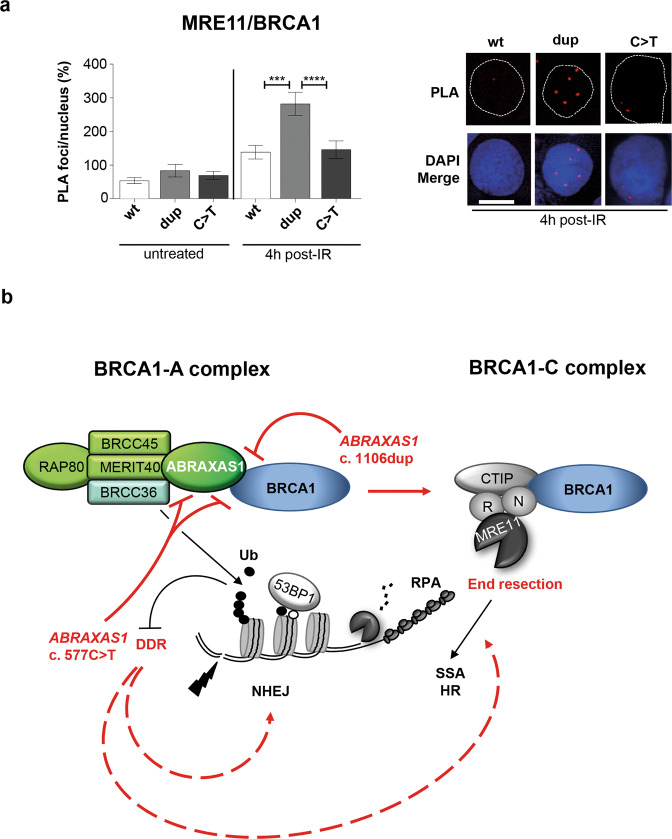
Fig. 7. Analysis of BRCA1-C complex formation and model for functional defects of ABRAXAS1 mutant proteins.
a Analysis of MRE11 and BRCA1 associations. LCL ABR-wt was transfected 24 h before IR with expression plasmids for wild-type (wt) or mutant ABRAXAS1 variants (dup, ABRAXAS1 c.1106dup; C>T, ABRAXAS1 c.577C>T). PLA was performed 4 h post IR. MRE11/BRCA1 PLA-foci were scored by automated quantification of 150 nuclei from three independent experiments and normalized to the mean for the external control transfected with empty vector, which was defined as 100% (0.2). Columns show mean values; n = 450; bars, SEM; Kruskal–Wallis test followed by two-tailed Mann–Whitney U test; **P < 0.01, ***P < 0.001, ****P < 0.0001. The right panel shows representative images of PLA-foci (red) 4 h post IR using primary antibodies anti-MRE11 (rabbit) and anti-BRCA1 (mouse) in DAPI-stained nuclei (blue). b Model for the findings revealing how the truncated ABRAXAS1 mutant proteins differentially deregulate DSBR. See main text for details. In brevity, ABRAXAS1 p.(Ser370Ilefs*2), encoded by ABRAXAS1 c.1106dup, cannot bind the C-terminal BRCT domains of BRCA1, therefore giving access to CTIP and MRE11-RAD50-NBS1. This causes a shift of BRCA1-A to BRCA1-C complex formation, which spurs end-processing and SSA at DSBs. ABRAXAS1 p.(Arg193*), encoded by ABRAXAS1 c.577C>T, additionally cannot bind BRCC36, which therefore cannot limit the DSB-induced ubiquitination cascade, entailing excess DNA damage signaling and unspecific DSBR-activities. Red color marks the impact of ABRAXAS1 mutant proteins on biochemical processes underlying the observed changes in DSBR.