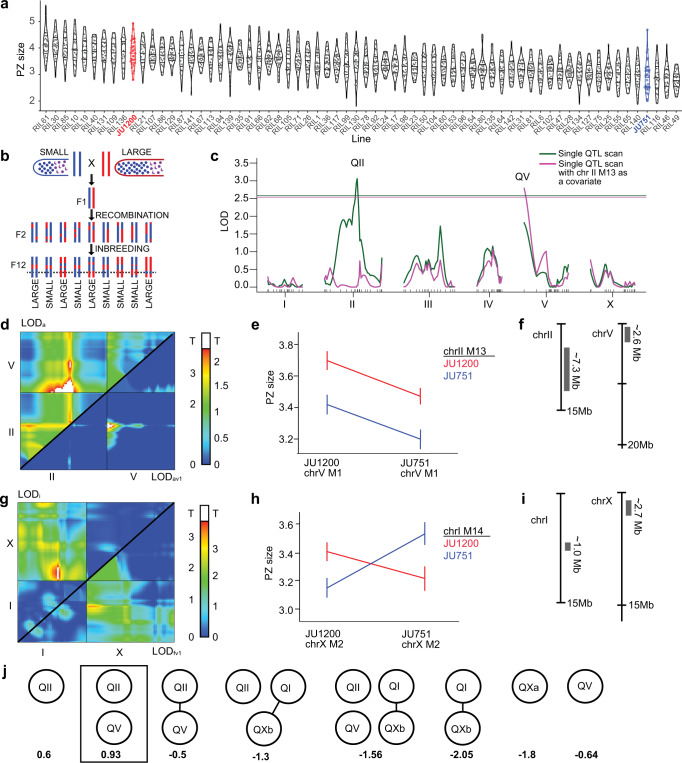
Fig. 3. Linkage mapping identifies multiple QTL explaining variation in PZ size.
a PZ size estimates (corrected) in F2 RILs (n = 70, assayed across six experimental blocks) and parental lines, JU1200 (red), and JU751 (blue) (see Supplementary Note 5 for details). b Generation and analysis of F2 RILs. Dotted line: hypothetical QTL. c Single-QTL scans identify two QTL (QII and QV). Horizontal lines: LOD threshold based on permutation tests. x-axis: genetic map for each chromosome. Green line: naïve scan results. Magenta line: results of a scan where marker M13 (peak, magenta curve) is included as a covariate. d Two-QTL scan results. Top half: LOD scores (LODa) comparing additive to null model. Bottom half: LOD scores comparing additive to single-QTL model (LODav1). LODav1 scores above threshold (white, determined by permutation testing) indicate improvement in fit over single-QTL model. e Interaction plot for markers in the peak in panel D, bottom half (chr V, M1; chr II, M14). f Approximate locations and sizes of the QTL on II and V. g Two-QTL scan allowing opposing effects. Top half: LOD scores (LODi) comparing a full two-QTL model to the additive model. Bottom half: LOD scores comparing the full model (allowing for interactions) with the single-QTL model (LODfv1). LODi scores above threshold (white) indicate evidence for an interaction, whereas LODfv1 scores above the threshold indicate an improvement in fit over the single-QTL model. h Interaction plot for markers in the significant peak in the top half of panel G (chr I, M14 and chr X, M2). i Approximate locations and sizes of candidate QTL on chromosomes I and X. j Representation of multi-QTL model selection. Nodes represent candidate QTL (QII = chrII@35 cM, QV = chr V@0 cM, QI = chrI@23 cM, QXa = chrX@16 cM, QXb = chrX@2.5 cM) and spokes represent interactions. Penalized LOD scores (below each model) above zero indicate better performance than the global null (zero QTL). See Methods, Supplementary Fig. 1, and Supplementary Note 6 for more detail. Y-axes in panels a, e, and h are scaled to μm2/1000 (0.1008 μm2/pixel). Data are provided as a Source Data file.