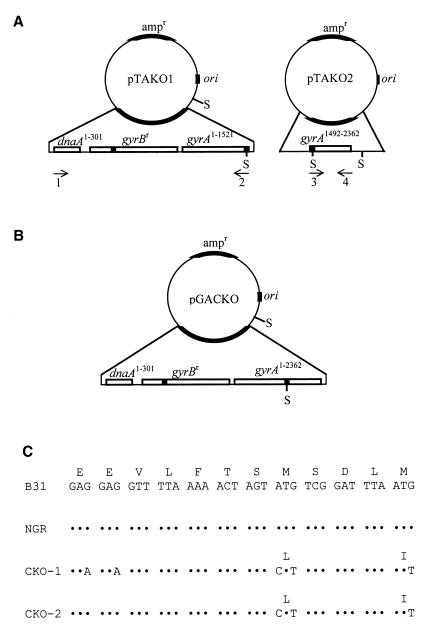
FIG. 1.
Strategy for gac gene disruption. (A) Plasmid maps of pTAKO1 and pTAKO2. Plasmid pTAKO1 contains the 5′ 301 bp of dnaA (dnaA1–301), all of gyrBr (dnaA and gyrB are divergently expressed), and the 5′ 1,521 bp of gyrA (gyrA1–1521). Plasmid pTAKO2 contains nucleotides 1492 to 2362 of gyrA (gyrA1492–2362). SpeI (S) restriction sites are shown. Arrows indicate oligonucleotide primers used in site-directed mutagenesis and PCR analysis; numbers below the arrows correspond to oligonucleotides listed in Table 1. Mutated sites in gyrB from B31-NGR and introduced mutations in gyrA are indicated by shaded boxes. (B) Plasmid map of pGACKO. The ∼800-bp SpeI fragment of plasmid pTAKO2 was cloned into the SpeI site of plasmid pTAKO1 to create plasmid pGACKO. (C) Nucleotide and amino acid sequences of gyrA from strains B31, B31-NGR, CKO-1, and CKO-2. Nucleotides 1474 to 1509 of gyrA, which encode amino acids 492 to 503 of GyrA (and amino acids 1 to 5 of Gac) are shown. Mutations are indicated with the corresponding amino acid change. Dots indicate nucleotide identity. Met 499 serves as the initiation codon for Gac, the naturally synthesized GyrA C-terminal domain.