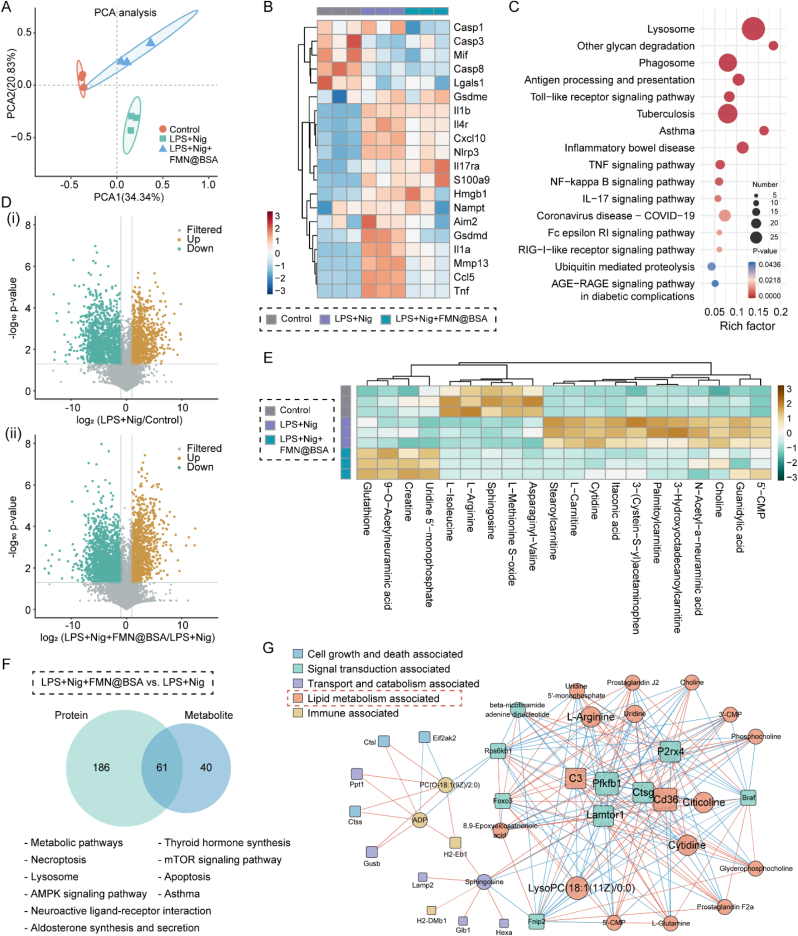
Fig. 5.
Combined proteomics/metabolomics analysis. A) Principal component analysis of proteomic data. PCA1 and PCA2 represent the largest source of variation. B) Heat map of differentially expressed proteins associated with pyroptosis in control, LPS+Nig, and LPS+Nig+FMN@BSA groups. C) Bubble plot of the KEGG pathway analysis enriched by differentially expressed proteins between LPS+Nig+FMN@BSA and LPS+Nig groups. The colors of the nodes reflect the p-values of the designated pathways, and the sizes of the nodes indicate the number of differentially expressed proteins enriched in the pathways. D) Volcano plots of differential metabolites in (ⅰ) pyroptotic and non-pyroptotic macrophages; (ⅱ) pyroptotic macrophages and treated ones. E) Heat map depicting the differential metabolites clustered between control, LPS+Nig, and LPS+Nig+FMN@BSA groups. F) Venn diagrams illustrating the number of metabolite (blue) and protein (green) pathways significantly changed in pyroptotic BMDMs after FMN@BSA NPs treatment, followed by a list of pathways that were regulated at both the protein and metabolite levels. G) Network analyses illustrating the key functional protein (squares) and metabolite (circles) nodes in pyroptotic BMDMs after FMN@BSA NPs treatment. Nodes were color-coded according to functional classification. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)