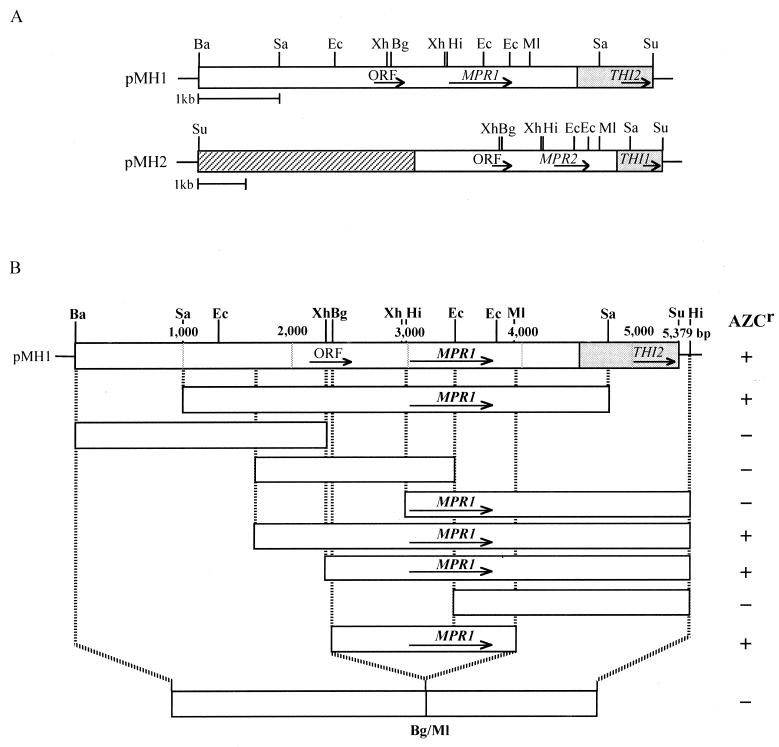
FIG. 1.
Restriction map of the cloned DNA fragment and deletion analysis to identify the region required for AZC resistance. The predicted size and transcriptional orientation of each deduced open reading frame (ORF) is shown by an arrow. Restriction enzymes: Ba, BamHI; Bg, BglII; Ec, EcoRI; Hi, HindIII; Ml, MluI; Sa, SacI; Su, Sau3AI; Xh, XhoI. (A) Restriction map of the cloned DNA fragment in pMH1 and pMH2. Two plasmids had the overlapping 5.4-kb Sau3AI insert (open box). The region in each plasmid matching the sequence on chromosome XIV (pMH1) or X (pMH2) of S. cerevisiae S288C is indicated by a shaded box. The hatched box in pMH2 represents the unknown, partially sequenced 4.6-kb fragment. (B) Analysis of MPR1 deletion mutants. Each DNA fragment was subcloned into pYES2, and the resultant plasmids were introduced into strain CKY2. AZC resistance of the Ura+ transformants was examined on SD agar plates containing AZC (0.3 mg/ml) after incubation at 30°C for 3 days. +, growth; −, no growth.