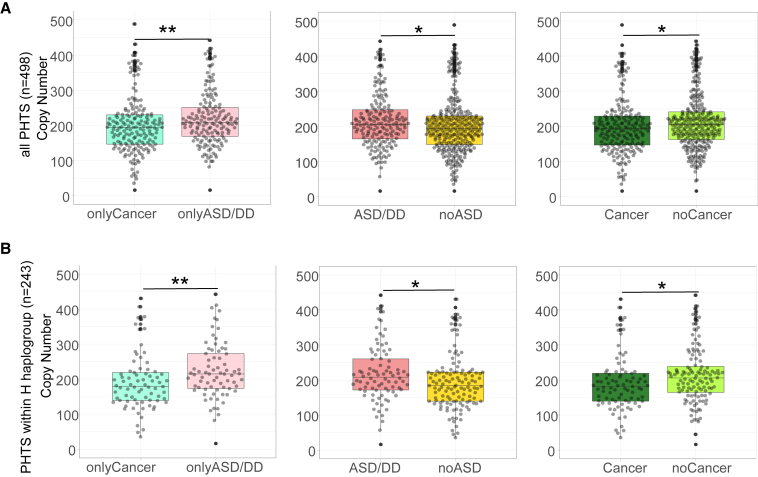
Figure 1.
Mitochondrial DNA copy number differences among different PHTS phenotype groups
(A) Beeswarm plots of copy number differences between different cancer/ASD phenotypes within all samples (n = 498). p values are calculated by the Mann-Whitney test.
(B) Beeswarm plots of copy number differences between different cancer/ASD phenotypes within the H haplogroup (n = 213). p values are calculated by the Mann-Whitney test.
The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles). Each dot represents one sample’s mtDNA CN value. The upper whisker extends from the hinge to the largest value no further than 1.5 ∗ inter-quartile range (IQR) from the hinge. The lower whisker extends from the hinge to the smallest value at most 1.5 ∗ IQR of the hinge. Data beyond the end of the whiskers are "outlying" points and are plotted individually.
∗∗0.001 < p value ≤ 0.01. ∗p value <0.05.