Fig. 1: TOGA utilizes intronic and intergenic alignments to detect orthologous gene loci.
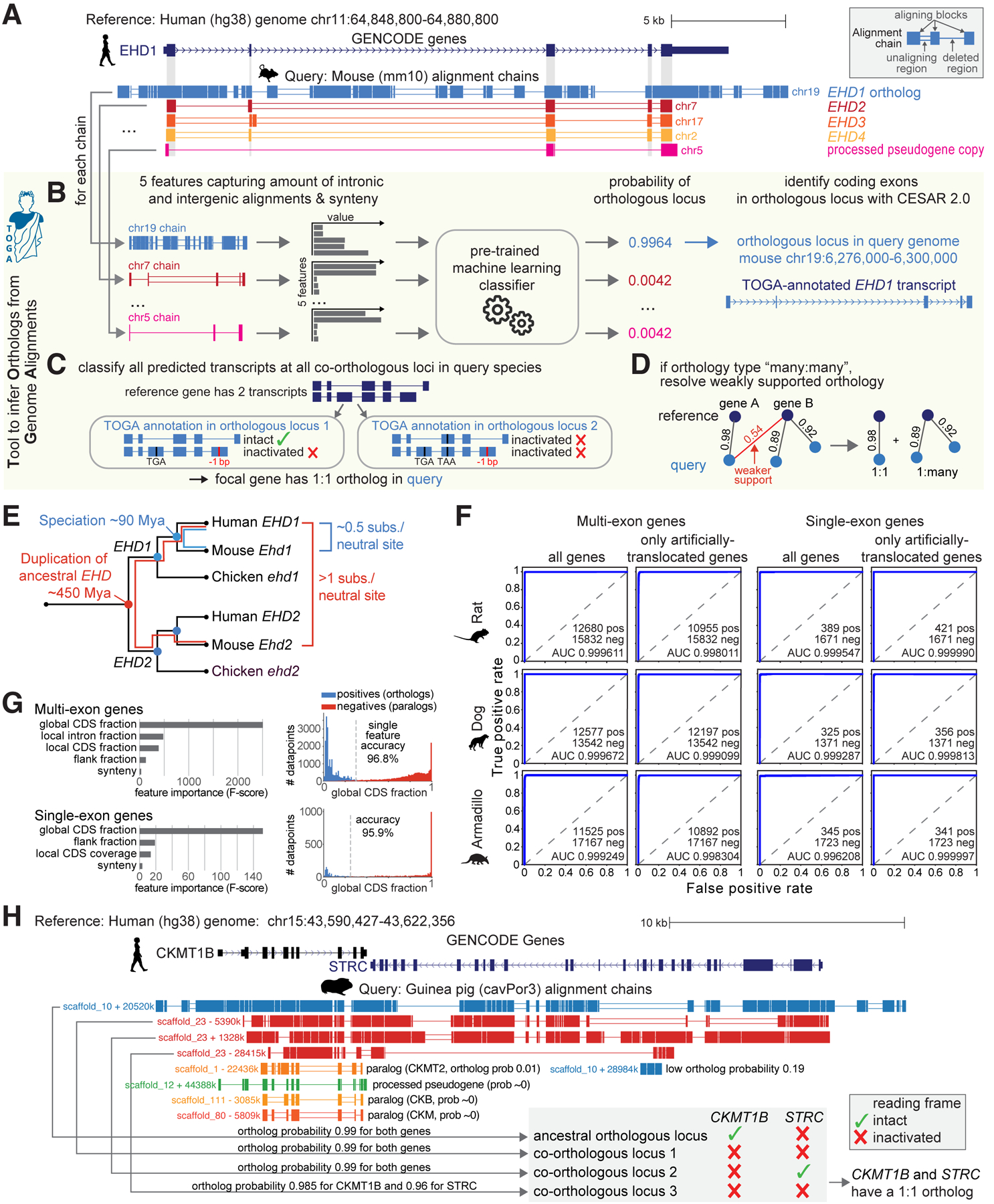
(A) UCSC genome browser view of the human EHD1 gene locus shows five alignment chains to mouse. Only the orthologous chr19 locus but not paralogous (chr7/17/2) and processed pseudogene (chr5) loci show intronic and intergenic alignments.
(B-D) Illustration of the TOGA pipeline steps that identify orthologous loci, annotate and classify transcripts, and resolve weak orthology connections.
(E) Evolutionary distance explains why only the orthologous EHD1 locus shows intronic and intergenic alignments.
(F) Orthology detection performance shown as Receiver Operating Characteristics curves for single- and multi-exon genes as well as for genes that lack synteny due to deliberately-introduced translocations.
(G) Feature importance for detecting orthologous genes and the distribution of the most important feature (“global CDS fraction”; proportion of coding exon alignments of all aligning chain blocks).
(H) Importance of detecting all orthologous loci and determining reading frame intactness. The human STRC and CKMT1B locus is quadruplicated in guinea pig (top four chains). TOGA correctly recognizes all four co-orthologous loci. Despite the quadruplication, TOGA finds that only one copy of each gene encodes an intact reading frame and correctly infers a 1:1 orthology relationship.
