Fig. 2: TOGA improves ortholog detection.
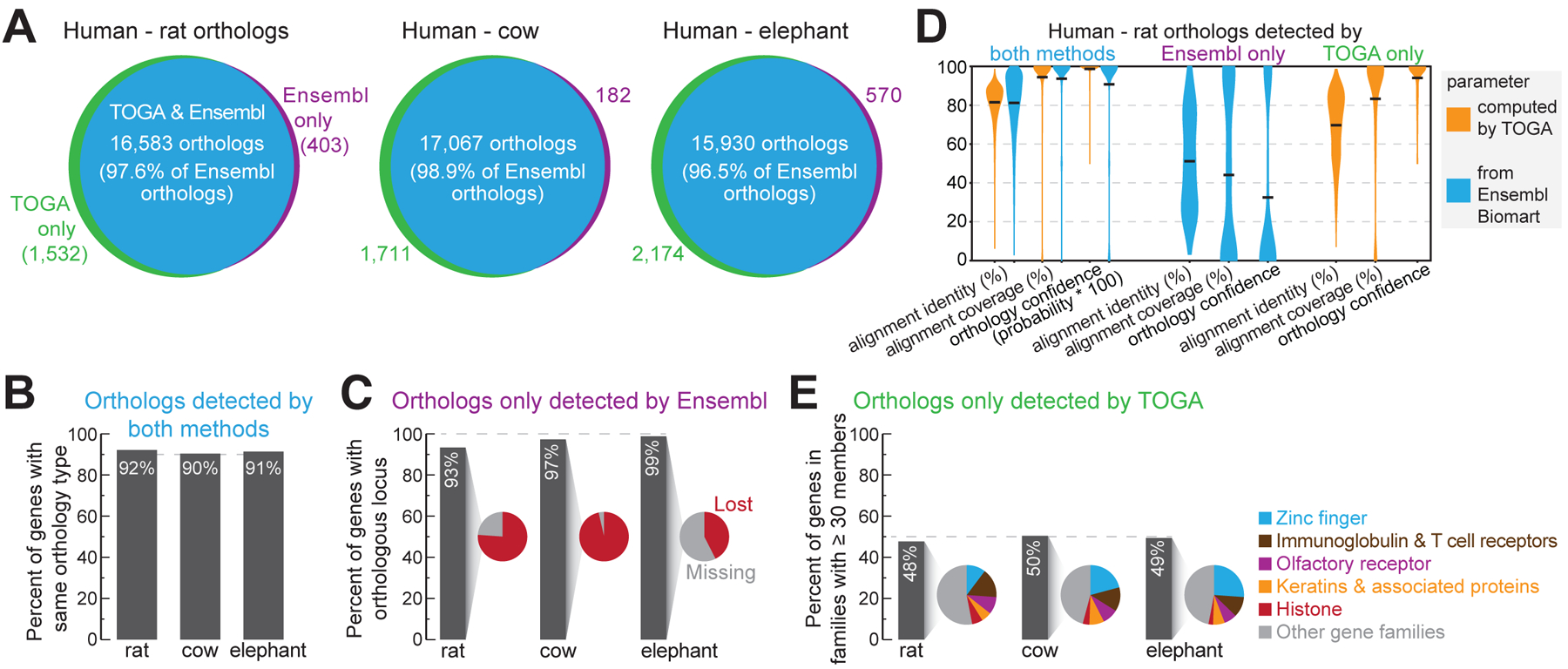
(A) Ortholog overlap between Ensembl Compara and TOGA.
(B) Percent of commonly-detected orthologs having the same orthology type.
(C) Percent of orthologs only detected by Ensembl, for which TOGA detects an orthologous locus but classifies the gene as lost or missing.
(D) Human-rat orthologs detected by both or only one method. Violin plots compare identity and coverage of coding region alignments and orthology confidence probabilities. Note that for orthologs only detected by TOGA, these features are not available on Ensembl Biomart, and vice versa. Horizontal black lines represent the mean.
(E) Percent of orthologs only detected by TOGA that belong to gene families with ≥30 members. Pie charts show the proportion of the most frequent gene families.
