Significance
Moths have served as historical models for animal communication studies. As their sex pheromone system is expected to be fine-tuned and under stabilizing selection, understanding its evolution has attracted interest for a long time. In the species Spodoptera littoralis and S. litura, the major component of the sex pheromone is an unusual compound, lacking in other species of the same genus. Building on the recent identification of a receptor narrowly tuned to this compound in S. littoralis, we investigated its evolution in the genus Spodoptera. We found that gene duplication followed by mutations that modified the binding pocket of the receptor led to the emergence of a new pheromone system in the common ancestor of S. littoralis and S. litura.
Keywords: pheromone receptor, evolution, ancestral gene resurrection
Abstract
Pheromone communication is an essential component of reproductive isolation in animals. As such, evolution of pheromone signaling can be linked to speciation. For example, the evolution of sex pheromones is thought to have played a major role in the diversification of moths. In the crop pests Spodoptera littoralis and S. litura, the major component of the sex pheromone blend is (Z,E)-9,11-tetradecadienyl acetate, which is lacking in other Spodoptera species. It indicates that a major shift occurred in their common ancestor. It has been shown recently in S. littoralis that this compound is detected with high specificity by an atypical pheromone receptor, named SlitOR5. Here, we studied its evolutionary history through functional characterization of receptors from different Spodoptera species. SlitOR5 orthologs in S. exigua and S. frugiperda exhibited a broad tuning to several pheromone compounds. We evidenced a duplication of OR5 in a common ancestor of S. littoralis and S. litura and found that in these two species, one duplicate is also broadly tuned while the other is specific to (Z,E)-9,11-tetradecadienyl acetate. By using ancestral gene resurrection, we confirmed that this narrow tuning evolved only in one of the two copies issued from the OR5 duplication. Finally, we identified eight amino acid positions in the binding pocket of these receptors whose evolution has been responsible for narrowing the response spectrum to a single ligand. The evolution of OR5 is a clear case of subfunctionalization that could have had a determinant impact in the speciation process in Spodoptera species.
Evolution of chemical communication between animals and their environment is crucial for adaptation to novel ecological niches as well as for the emergence of new species. It is especially true when intraspecific chemical signals, called pheromones, are used for finding and selecting mating partners and prevent closely related species from interbreeding, thus conferring reproductive isolation. Changes in such prezygotic isolation mechanisms within a population can create reproductive barriers, which can be a driver for speciation (1, 2). Among the best-known examples of pheromone communication is the sex pheromone system of moths. In these insects, reproductive success largely depends on mate recognition through the detection by males of specific bouquets of volatile compounds emitted by females. These compounds are mostly long-chain acetates, alcohols, or aldehydes synthesized de novo from fatty acids. Moth sex pheromone blends usually contain one major component and one or more minor components in a precise ratio, and blends of closely related species typically differ only in a few minor components or in the ratio between the components (3). Male antennae bear olfactory sensory neurons (OSNs) that detect these components with various sensitivities and specificities, allowing a precise encoding of the blend composition. This encoding underlies male preference toward a given pheromone.
At the molecular level, this detection is mediated by pheromone receptors (PRs) belonging to the insect odorant receptor (OR) family. Like other insect ORs, PRs have seven transmembrane domains and assemble in a heteromultimer composed of a ligand-selective receptor and the universal coreceptor Orco, which together act as a ligand-gated ion channel (4–6). Moth PRs are highly––sometimes specifically––expressed in male antennae and tuned to one or a few structurally related pheromone compounds. They allow males to detect pheromone compounds released by conspecific females but also by sympatric species, which eventually prevents cross-species mating and reinforces reproductive isolation (7). PRs can also be expressed at lower levels in females as well as in larvae, although the role of sex pheromone sensing outside males remains unclear (8). How pheromone detection and male preferences, which are expected to be fine-tuned and under a strong stabilizing selection, evolve to allow the emergence of novel pheromone communication channels is a fascinating yet largely unresolved question (1, 9).
From an evolutionary point of view, insect Or genes follow the classical birth-and-death model of evolution of multigene families, characterized by high rates of tandem duplications, ultimately leading to functional divergence and the emergence of novel functions, as well as gene deletions or pseudogenization events (10, 11). Moth PRs make no exception and have a highly dynamic evolutionary history (12), although the implication of gene gains and loss in the evolution of PR response spectra has not been investigated per se. Single adaptive mutations explaining differences of ligand specificity between orthologous PRs from pairs of closely related species have been documented (13–15), but evolutionary studies across a broader range of species are lacking.
Moths of the genus Spodoptera (family Noctuidae) are good models for exploring the question of the evolution of pheromone detection. Sex pheromones have been characterized in a dozen of Spodoptera species (16) and consist of mixtures of 14-carbon acetates in a specific ratio, with (Z)-9-tetradecenyl acetate being the major component of the blend in most species. However, in the two sibling species S. littoralis and S. litura, the major component of the sex pheromone is (Z,E)-9,11-tetradecadienyl acetate (hereafter referred to as (Z,E)-9,11-14:OAc), which is lacking or present only in minute amounts in other species (SI Appendix, Fig. S1). Detection of this compound is necessary and sufficient to trigger male attraction (17, 18). This means that a major change in the pheromone communication system occurred in a common ancestor of S. littoralis and S. litura. We recently discovered in S. littoralis a receptor tuned to (Z,E)-9,11-14:OAc, named OR5 (17). Interestingly, this receptor is distantly related to other moth PRs, suggesting independent origins. Building on this finding, we aimed at investigating the evolutionary trajectory of OR5 in the genus Spodoptera, in order to shed light on events that allowed the emergence of a new pheromone communication channel in the common ancestor of S. littoralis and S. litura.
Results
The Or5 Gene Duplicated in an Ancestor of S. littoralis and S. litura.
Our first objective was to rebuild the evolutionary history of Or genes in the genus Spodoptera and identify duplications and losses that may have occurred in a common ancestor of S. littoralis and S. litura. A repertoire of 74 Or genes has recently been annotated in the reference genome of S. littoralis (19). Here, we used all chromosome-level genome assemblies publicly available on NCBI to annotate de novo 70 Ors in S. exigua, and to reannotate 71 Ors in S. frugiperda––i.e., one more than in another genome assembly (20)––and 75 Ors in S. litura––two more than previously identified (21). Phylogenetic analysis revealed that these Ors group into 72 distinct paralogous lineages, which evolved each from a gene that was present in the common ancestor of the four Spodoptera species (Fig. 1A and SI Appendix, Fig. S2). Or gains and losses occurred in seven of these paralogous lineages, including three duplications in a common ancestor of S. littoralis and S. litura. The receptor tuned to (Z,E)-9,11-14:OAc in S. littoralis, SlitOR5, is encoded by one of these duplicated genes. We identified SlitOr5 orthologs in all the three S. exigua (Sexi), S. frugiperda (Sfru), and S. litura (Slitu) genomes, and also found a SlitOr5 paralog positioned in tandem in the genomes of S. littoralis and S. litura only, named Or75 (Fig. 1B). The phylogenetic analysis showed that Or5 and Or75 paralogs arose from a recent duplication that occurred in a common ancestor of S. littoralis and S. litura (Fig. 1C). The OR proteins encoded by the four Or5 and the two Or75 genes exhibit a ~70% mean amino acid sequence identity, except SlitOR5/SlituOR5 and SlitOR75/SlituOR75 pairs that share 94% identity (Fig. 1D).
Fig. 1.
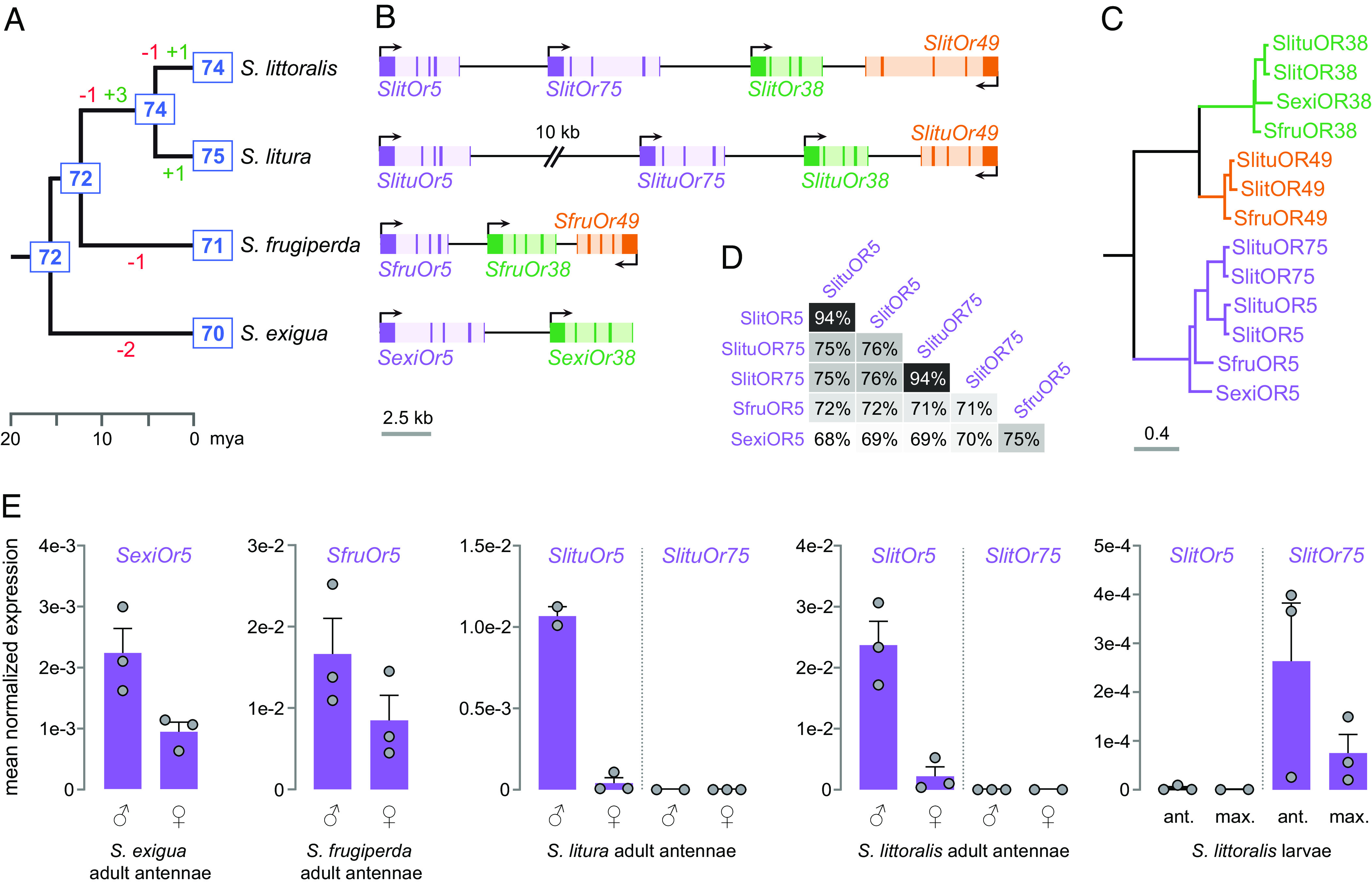
Or5 orthologs and paralogs in four Spodoptera species. (A) Evolution of the number of Or genes (blue boxes) in the genus Spodoptera as determined by the phylogenetic analysis (SI Appendix, Fig. S1). Gene gains are shown in green and losses in red. Phylogenetic relationships and divergence times are from ref. 22. (B) Synteny of Or5 orthologs and paralogs in the four species. Exons are shown with dark colors, introns with light colors. Arrows indicate the 5′ end of the ORF and the direction of transcription. (C) Phylogeny of ORs encoded by genes shown in B. The scale bar shows the expected number of amino acid substitutions per site. (D) Identity matrix between amino acid sequences encoded by Or5 and Or75 genes. (E) Expression level of Or5 and Or75 genes in olfactory organs of the four species as determined by RT-qPCR. Reference genes were ATPs for S. exigua, Actin for S. frugiperda, and Rpl13 for S. litura and S. littoralis.
As moth sex PRs are generally overexpressed in male antennae (17), we then sought to compare expression levels of Or5 and Or75 in adult antennae of the four Spodoptera species. In S. exigua and S. frugiperda, SexiOr5 and SfruOr5 were expressed in both male and female antennae with a twofold enrichment in males (Fig. 1E). By contrast, SlitOr5 and SlituOr5 presented a clear male-biased expression in S. littoralis and S. litura antennae (10-fold and 26-fold enrichment compared to females, respectively). Expression of the SlitOr75 and SlituOr75 duplicates was nearly undetectable in both sexes. We tested whether Or75 may be expressed in olfactory organs of S. littoralis at the larval stage and found a moderate yet consistent expression of SlitOr75, but not SlitOr5, in antennae and maxillae of fourth-instar larvae. Altogether, these results indicate that the expression level of Or5 changed according to sex during Spodoptera spp. evolution. Following Or5 duplication in a common ancestor of S. littoralis and S. litura, one copy became highly expressed in adult male antennae.
Response Spectra of OR5 and OR75 Differ between the Four Spodoptera Species.
To investigate how the function of receptors encoded by Or5 and Or75 genes evolved in the genus Spodoptera, we verified whether they were all able to bind pheromone compounds and compared their tuning breadths. To do so, we used heterologous expression in at1 OSNs of Drosophila (23), a class of olfactory neurons housed in antennal trichoid sensilla and used by flies to detect the antiaphrodisiac pheromone cis-vaccenyl acetate (23). This expression system has proved successful for studying SlitOR5 (17). Single-sensillum recordings were performed to measure the response of transformed at1 OSNs to a large panel of moth pheromone compounds including all those identified in the sex pheromone blends of Spodoptera species (SI Appendix, Fig. S1). First, we investigated the response profiles of OR5 orthologs from S. exigua and S. frugiperda, whose females do not produce (Z,E)-9,11-14:OAc in their pheromone blends. We found that OSNs expressing SexiOR5 responded significantly to 10 compounds, and the highest activation was recorded in response to (Z,E)-9,11-14:OAc (Fig. 2A). OSNs expressing SfruOR5 also responded significantly to 10 compounds, most of them being the same as for SexiOR5, with the highest response recorded for (E)-11-14:OAc, a minor component of the S. frugiperda pheromone. Dose–response curves confirmed that SfruOR5 and SexiOR5 bound similar compounds, yet with different sensitivities (Fig. 2B). None of the ligands identified belongs to the pheromone blends of these two species (SI Appendix, Fig. S1). Second, we found that the function of OR5 orthologs was conserved between S. littoralis and S. litura, who share the same major pheromone component (Z,E)-9,11-14:OAc. Indeed, only this compound elicited strong responses of OSNs expressing SlitOR5 or SlituOR5, although a significant yet minor response to (Z,Z)-9,11-14:OAc was also measured for SlituOR5 (Fig. 2A). Dose–response analyses revealed that SlitOR5 and SlituOR5 had similar detection thresholds (Fig. 2B).
Fig. 2.
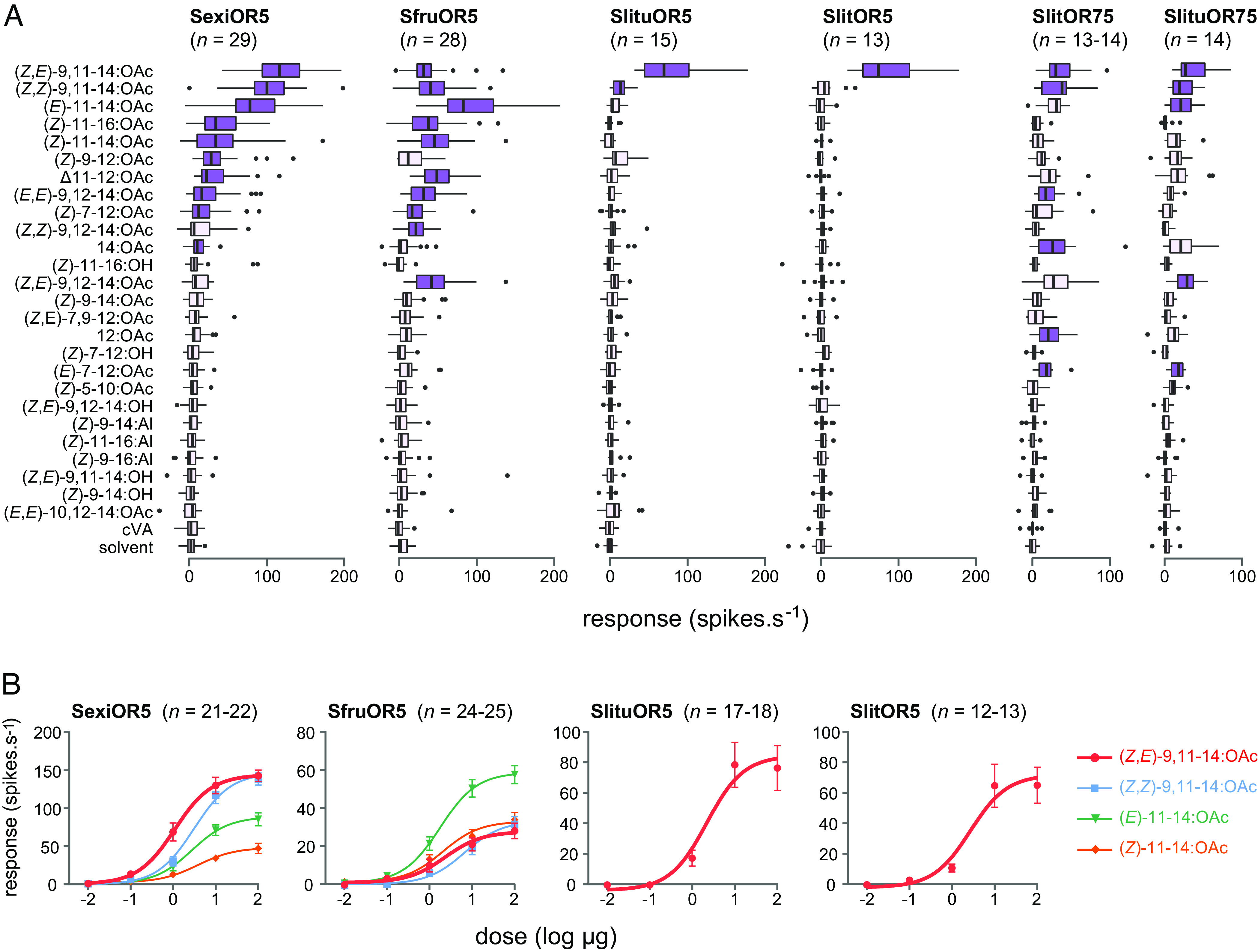
Tuning breadth of OR5 orthologs and paralogs in Spodoptera species. (A) Action potential frequency recorded in Drosophila at1 OSNs expressing Or5 and Or75 genes of the four species after stimulation with 26 sex pheromone compounds (10 μg loaded in the stimulus cartridge). Boxes show the first and third quartiles, whispers show the distribution, and dots show outliers. Dark purple shows responses significantly different from the response to solvent (P < 0.05, one-way ANOVA followed by a Dunnett’s post-hoc test). (B) Dose–response curves of the four OR5 orthologs. For SexiOR5 and SfruOR5, four compounds were tested. For SlituOR5 and SlitOR5, which were specific to (Z,E)-9,11-14:OAc, only this compound was tested.
Additionally, expression in Xenopus laevis oocytes and two-electrode voltage-clamp recordings were used to confirm the response spectra of SlituOR5, SfruOR5, and SexiOR5. The two heterologous expression systems revealed highly similar response profiles (SI Appendix, Fig. S3), hereby confirming that SlitOR5 and SlituOR5 are narrowly tuned to (Z,E)-9,11-14:OAc, whereas SfruOR5 and SexiOR5 are more broadly tuned and can bind several pheromone compounds, including (Z,E)-9,11-14:OAc.
Since we evidenced a duplication of Or5 in both S. littoralis and S. litura, we next investigated the function of the Or75 duplicates. OSNs expressing SlitOR75 or SlituOR75 exhibited low responses to (Z,E)-9,11-14:OAc as well as four and five other compounds from the panel, respectively (Fig. 2A). Taken together, these results led us to hypothesize that the ancestral Spodoptera OR5 was a broadly tuned pheromone receptor able to detect (Z,E)-9,11-14:OAc among other molecules, and that following duplication in a common ancestor of S. littoralis and S. litura, one copy retained a broad tuning while the other evolved a narrow tuning to (Z,E)-9,11-14:OAc.
Resurrected Ancestral ORs Are Functional and Have Similar Tuning Breadths Than Those of Their Descendants.
One way to test these hypotheses is to study the evolutionary trajectory of genes of interest by testing the function of ancient genes, a process known as ancestral gene resurrection (24–26). Here, we resurrected three ancestral receptors (further referred to as AncORs) located at three critical nodes in the phylogeny (Fig. 3A). One ancestor was located before the Or5/Or75 duplication (AncOR5_75), the two others after the duplication but before the split between S. littoralis and S. litura (AncOR5 and AncOR75). We first reconstructed their amino acid sequences using a maximum likelihood and Bayesian framework. For each amino acid position, a posterior probability was computed. Overall, the amino acids inferred by the ancestral reconstruction were supported by high posterior probabilities (SI Appendix, Fig. S4). The reconstructed sequences of AncOR5 and AncOR75 exhibited 96 to 97% identity with the extant receptors of S. littoralis and S. litura (Fig. 3B and SI Appendix, Fig. S5). AncOR5_75 exhibited 87% identity with AncOR5 (which corresponds to 50 amino acid difference), 90% identity with AncOR75 (38 amino acid difference), and 84 to 87% identity with the extant receptors of S. littoralis and S. litura.
Fig. 3.
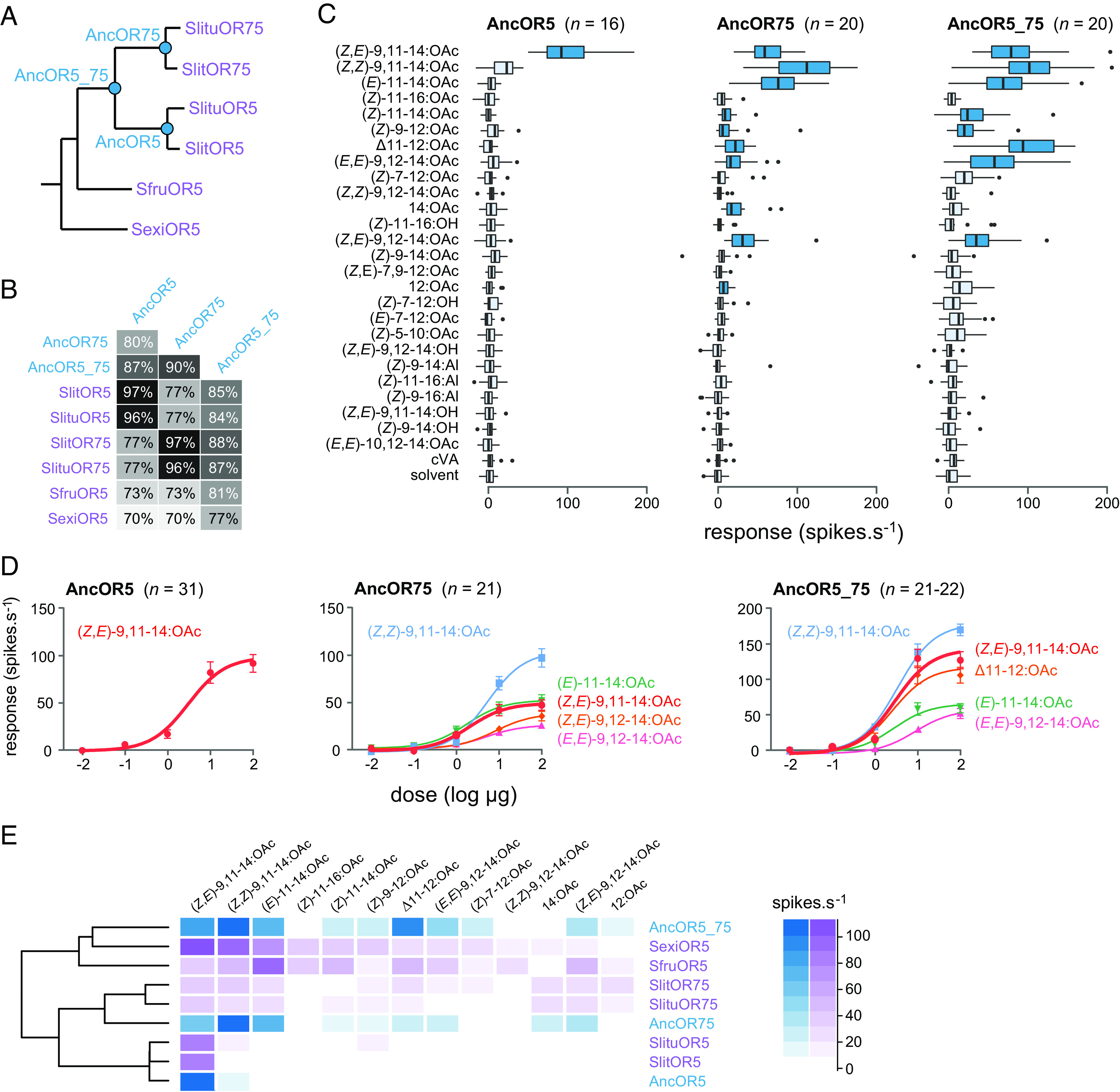
Tuning breadth of resurrected ancestral ORs before and after the Or5 duplication in the ancestor of S. littoralis and S. litura. (A) Phylogeny of OR5 orthologs and paralogs (purple). The ancestral sequences that were resurrected are indicated on the nodes (blue). (B) Amino acid sequence identity matrix between the three ancestral and the six extant ORs. (C) Action potential frequency recorded in Drosophila at1 OSNs expressing Or5/Or75 ancestors after stimulation with 26 sex pheromone compounds (10 μg loaded in the stimulus cartridge). Boxes show the first and third quartiles, whispers show the distribution, and dots show outliers. Dark blue shows responses significantly different from the response to solvent (P < 0.05, one-way ANOVA followed by a Dunnett’s post-hoc test). (D) Dose–response curves of the three ancestral ORs. (E) Heatmap summarizing the tuning breadth of OSNs expressing ancestral (blue) and extant Or5/Or75 genes (purple). Receptors were clustered using Manhattan distances (dendrogram on the left).
The reconstructed sequences were then synthesized in vitro and the AncORs were resurrected by expressing them in Drosophila OSNs, in order to determine their tuning breadth. All the three AncORs were functional. AncOR5-expressing OSNs responded specifically to (Z,E)-9,11-14:OAc, whereas OSNs expressing AncOR75 and AncOR5_75 yielded significant responses to 10 and 8 pheromone compounds, respectively (Fig. 3C). These last two ancestors bound (Z,E)-9,11-14:OAc with the same activation threshold as AncOR5 but they were equally sensitive to other compounds, with (Z,Z)-9,11-14:OAc as their best ligand (Fig. 3D). In summary, the response spectra of AncOR75 and AncOR5_75 matched those of the extant OR5 and OR75 receptors in the four Spodoptera species except SlitOR5 and SlituOR5 (Fig. 3E), thus supporting our hypotheses that OR5 was ancestrally able to bind several pheromone compounds including (Z,E)-9,11-14:OAc and that following duplication, one copy became narrowly tuned to this compound.
Receptor Tuning Is Determined by Multiple Amino Acid Positions.
In order to decipher the molecular basis of the narrowing in tuning between AncOR5_75 and AncOR5, we modeled their 3D structure with AlphaFold2 and determined their putative ligand binding pockets, as well as the (Z,E)-9,11-14:OAc pose in AncOR5_75 (Fig. 4A). Nine different poses were obtained when docking (Z,E)-9,11-14:OAc to the modeled structure of AncOR5_75. Residues located within 5 Å from all the nine poses of the ligand were considered as potential interacting residues. Out of these 28 amino acid positions that could possibly interact with the pheromone compound, eight were particularly interesting because they differed between AncOR5_75 and AncOR5 (Fig. 4B and SI Appendix, Fig. S5). To verify whether these amino acid changes were indeed responsible for the change in tuning breadth, we retraced the potential evolutionary trajectory by substituting the eight AncOR5_75 putative binding pocket residues by homologous residues of AncOR5. A total of 12 chimeric mutants were obtained by site-directed mutagenesis, including the eight different single-site mutants (M76V, V79L, T140V, A143T, F169Y, A204V, I275V, and T276S), two double mutants (V79L_A143T and I275V_T276S), one triple mutant (V79L_A143T_A204V), and one mutant carrying the eight mutations named AncOR5_75_mut8x. For double and triple mutants, residues in close proximity to each other were selected based on the predicted 3D structure.
Fig. 4.
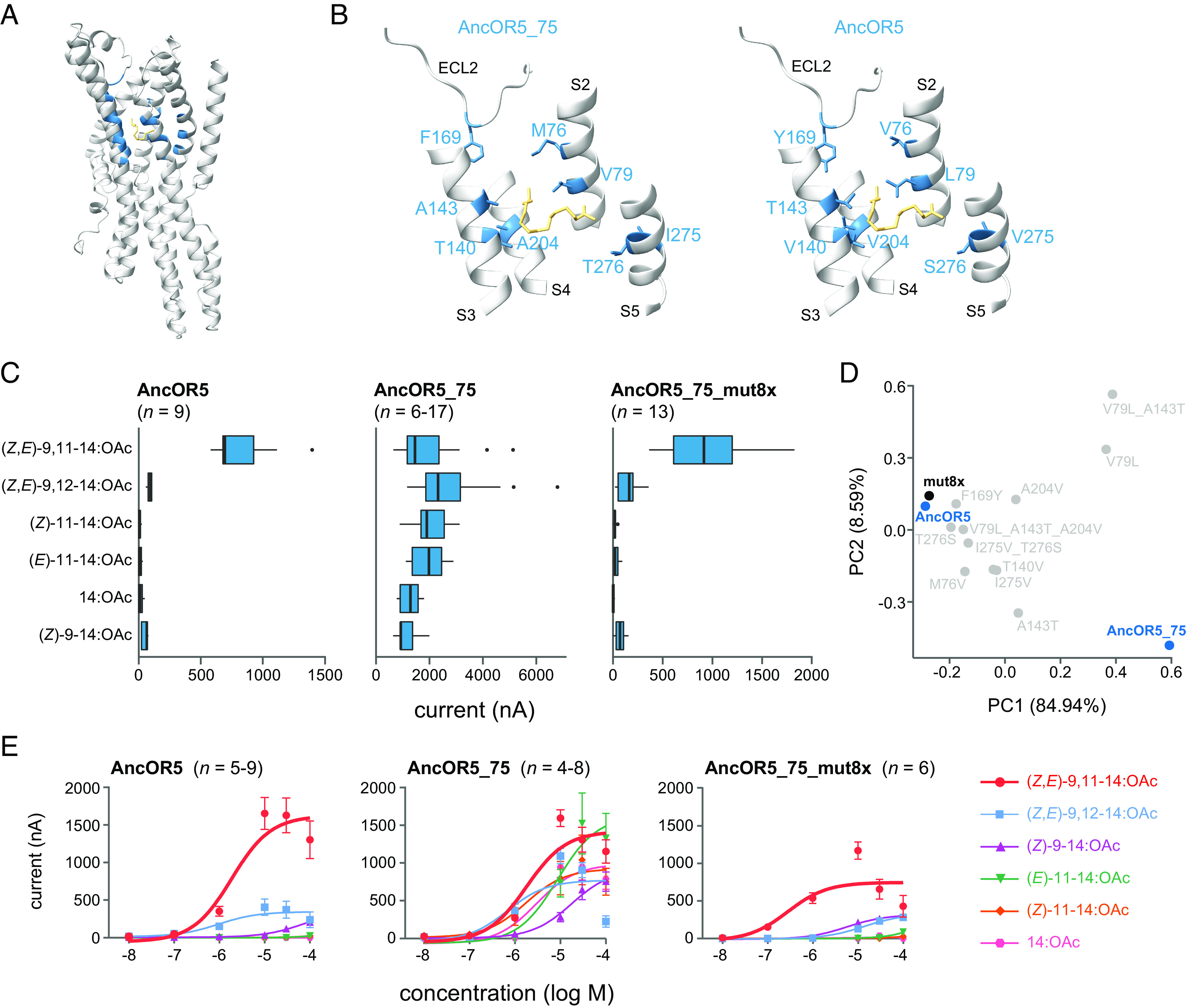
Structural basis for the evolution of OR5 tuning breadth. (A) 3D structure of AncOR5_75 as predicted by AlphaFold2. The ligand (Z,E)-9,11-14:OAc is shown in yellow. The 28 amino acid positions predicted to interact with the ligand (distance <5 Å) are shown in blue. (B) Binding pockets of AncOR5_75 and AncOR5 showing the eight amino acids differing between the two ancestors. S2, 3, 4, and 5: transmembrane helices; ECL2: extracellular loop 2. (C) Inward current measured in Xenopus oocytes coexpressing AncOR5, AncOR5_75, or AncOR5_75_mut8x and SlitOrco after stimulation with a panel of six components from the S. littoralis sex pheromone (10−5 M solution). Responses to the negative control (1× Ringer’s buffer containing 0.01% DMSO) were 0. (D) Principal component analysis based on the responses of AncOR5, AncOR5_75, and its related mutants. (E) Dose–response curves of AncOR5, AncOR5_75, and AncOR5_75_mut8x expressed in Xenopus oocytes.
All mutants as well as AncOR5_75 and AncOR5 were individually coexpressed with SlitOrco in Xenopus oocytes, and we recorded their electrophysiological responses to 6 pheromone compounds of S. littoralis. AncOR5_75 responded to all the six compounds, while AncOR5 specifically responded to (Z,E)-9,11-14:OAc (Fig. 4C), similarly to what we observed when expressed in Drosophila OSNs. Interestingly, AncOR5_75_mut8x mirrored the response profile of AncOR5 in terms of specificity (Fig. 4 C and D and SI Appendix, Fig. S6), showing that the eight positions are indeed key for the receptor tuning.
Dose–response experiments were performed to further assess the sensitivity of AncOR5_75_mut8x, AncOR5_75, and AncOR5 to the different pheromones. AncOR5_75 showed a broad tuning with comparable sensitivities to most ligands, whereas AncOR5 and AncOR5_75_mut8x were more sensitive to (Z,E)-9,11-14:OAc with EC50 values of 1.1 µM and 0.4 µM, respectively (Fig. 4E). All the individual mutations or double/triple combinations showed an effect on AncOR5_75 tuning and most of them reduced its tuning breadth, but none of these mutations recapitulated the tuning of AncOR5 and AncOR5_75_mut8x (Fig. 4D). Notably, the triple-mutant V79L_A143T_A204V exhibited a narrow tuning to the stereoisomer (Z,E)-9,12-14:OAc (SI Appendix, Fig. S7). Altogether, this suggests that multiple mutations in the binding pocket have been involved in the tuning of OR5 toward (Z,E)-9,11-14:OAc.
Discussion
In this study, we have identified and functionally characterized five ORs from Spodoptera species that all clustered in the unique novel pheromone receptor clade containing SlitOR5. They all bound sex pheromone compounds, either from their own pheromone blend or from that of other moth species. We thus further confirm the previous findings that moth PRs tuned to type I pheromone compounds (i.e., straight-chain hydrocarbons of 10 to 18 carbons carrying a terminal acetate, alcohol, or aldehyde function) arose at least twice during lepidopteran OR evolution (17, 27, 28). Spodoptera OR5 orthologs presented different tuning breadths and expression patterns. OR5 from the sister species S. littoralis and S. litura showed a male-biased expression and were specifically tuned to (Z,E)-9,11-14:OAc, their major sex pheromone component. In the distantly related species S. exigua and S. frugiperda that do not use (Z,E)-9,11-14:OAc as a pheromone component, OR5 presented broader response spectra and did not show a strong sex-biased expression. Interestingly, they seem to serve different functions in these species. SexiOR5 was tuned to several heterospecific compounds, i.e., which are not present in the S. exigua sex pheromone blend, with (Z,E)-9,11-14:OAc being the best ligand. S. exigua lives in sympatry with S. littoralis and S. litura (29), and it has been shown that S. littoralis and S. litura males can detect the major pheromone component of S. exigua (Z)-9-14:OH, which reduces catches in traps baited with their own sex pheromones (30, 31). This behavior could have been selected to maintain reproductive isolation between sympatric species. Similarly, S. exigua males are able to detect (Z,E)-9,11-14:OAc, which decreases trap catches (31). SexiOR5 is most probably the receptor responsible for this antagonistic behavior. In S. frugiperda, SfruOR5 also bound several components including (Z,E)-9,11-14:OAc) and its response spectrum closely resembled that of an OSN population in S. frugiperda male antennae (32). The best SfruOR5 ligands were either (E)-11-14:OAc or (Z)-11-14:OAc, depending on the expression system. These compounds have been found at minute amounts in the pheromone glands of S. frugiperda females and their presence at 1% in synthetic pheromone blends strongly increased the proportion of males approaching the source in wind tunnel experiments (32). SfruOR5 may thus play an important role in pheromone communication by detecting minor components with a synergistic effect. In addition to the detection of heterospecific compounds (in S. exigua) and minor pheromone components (in S. frugiperda), we identified another potential function for OR5 homologs. In S. littoralis, Or5 is mostly expressed in adult male antennae and is necessary for male attraction toward the pheromone, whereas the duplicate Or75 is expressed in larvae chemosensory organs. Expression of this latter receptor could explain how S. littoralis larvae can smell the female sex pheromone (33). This finding points toward the importance of considering the evolution of receptor expression and not only the evolution of response spectra. Further studies are needed to understand pathways regulating Or expression and how they can evolve in moths (34).
Regarding the shift in specificity of OR5 in S. littoralis and S. litura compared to OR75 paralogs and to S. frugiperda and S. exigua orthologs, the ancestral resurrection results suggest a typical case of subfunctionalization. Before the S. littoralis/S. litura split, AncOR5_75 likely acted as a broadly tuned receptor, already able to detect (Z,E)-9,11-14:OAc. According to the asymmetric tracking hypothesis (35, 36), the broad tuning of this OR may have enabled males to respond to unusual pheromone components, like (Z,E)-9,11-14:OAc, emitted by some females carrying a mutation in the pheromone biosynthesis pathway. However, after the OR5/OR75 duplication, OR5 acquired a drastically narrowed response spectrum to specifically detect (Z,E)-9,11-14:OAc, while its duplicate, OR75, retained a broad tuning. This suggests a rapid acquisition of different odor affinities after duplication, an essential step to avoid gene loss and eliminate redundancy. What remains puzzling is that, despite the short evolutionary time, we showed here that multiple amino acid substitutions seem to be necessary for the shift of ligand specificity between the ancestors and the actual OR5 receptors. This is an original finding since previous studies identified that only one or two amino acid substitutions are sufficient for a shift of function in other ecologically important receptors (13–15, 37–41). Our study clearly demonstrated that in the case of OR5, one amino acid substitution does not have a drastic impact on ligand specificity. However, the precise evolutionary trajectory that led to the shift of function and whether all the 8 mutations identified are necessary remain unknown. Further studies to narrow down the key residues should be conducted in the future. The analysis of sequence data from different populations of S. littoralis and S. litura could also help to identify the most critical residues in the OR5 fitness landscape.
Resurrection of ancestral ORs played a pivotal role first in deciphering the evolutionary trajectory of the OR5/75 clade and, second, in decreasing the number of putative amino acid candidates to test for functional analysis. To further narrow down the number of residues, we combined ancestral resurrection with 3D modeling. The combination of both approaches proved useful as only eight amino acids were ultimately pinpointed. A similar combined approach was conducted on other genes and permitted to identify residues that are critical for the protein function in chemoreceptors (42) and also in metabolic enzymes (43, 44). With the exponential rise of sequence data, mandatory for ancestral reconstruction reliability, the progress in molecular modeling with AlphaFold2 and other highly efficient modeling tools (45), as well as the easiness of synthesis of nonexisting sequences, all prerequisites are available to implement such an integrative functional assessment method (24).
Material and Methods
Annotation of Or Genes and Phylogeny.
Or genes were annotated by aligning a set of 485 full-length amino acid sequences of Noctuidae ORs with genome assemblies of Spodoptera species using Exonerate v2.4.0 (46) as implemented in Galaxy (47), with a score threshold of 500 and only the three best results reported. To avoid overestimating gene losses because of genes potentially missing in a given assembly, we used all chromosome-level assemblies available at NCBI for S. exigua (WH-S isolate, NJAU_Sexi_v1, GCA_011316535.1; QAU_Sexi_v1, GCA_022315195.1; PGI_SPEXI_v6, GCA_902829305.4), S. frugiperda (ZJU_Sfru_1.1 Zhejiang isolate, GCA_011064685.2; ASM1297921v2, GCA_012979215.2; AGI-APGP_CSIRO_Sfru_2.0, GCA_023101765.3), and S. litura (ASM270686v3Ishihara strain, GCA_002706865.3). Redundant sequences were eliminated using CD-HIT EST v1.3 (48) with a similarity threshold of 0.98. For the phylogenetic analysis, OR amino acid sequences were aligned with Clustal Omega v1.2.2 (49) and the tree was built with PhyML v3.0 (50) Branch support was estimated using a SH-like approximate likelihood ratio-test (51).
Insect Rearing and Quantitative Real-Time PCR.
S. exigua and S. litura were bought from the Jiyuan Baiyun Industry Company (China). S. frugiperda were collected from a corn field in Yunnan province (China) in March 2019 and maintained in the lab on an artificial diet at 26 °C, 60% relative humidity, and under a 14 h:10 h light:dark cycle. S. littoralis originated from a laboratory colony reared on a semi-artificial diet (22 °C, 60% relative humidity, 16 h light:8 h dark). For all species, males and females were sexed as pupae and further reared separately.
Total RNA was extracted from three independent replicates of antennae (except for S. litura males, two replicates) from 2- to 3-d-old virgin males and females of S. exigua (50 pairs), S. frugiperda (40 pairs), S. litura (30 pairs), and S. littoralis (15 pairs) as well as from three replicates of 160 pairs of antennae and maxillae of fourth-instar S. littoralis larvae using TRIzol™ Reagent (Thermo Fisher Scientific, Waltham, MA, USA), including a DNase I treatment. cDNA was synthesized from 1 µg total RNA using the RevertAid First-Strand cDNA synthesis kit or SuperScript™ II reverse transcriptase (Thermo Fisher Scientific). Gene-specific primers (SI Appendix, Table S1) were designed using Primer3 (http://primer3.ut.ee) or AmplifX (http://inp.univ-amu.fr/). All reactions were performed in triplicate for each biological replicate. For each gene, a negative control without cDNA and a fivefold dilution series of pooled cDNAs from all conditions were included. For S. exigua and S. frugiperda, the qPCR mix contained 10 µL GoTaq® qPCR Master Mix (Promega, Madison, WI, USA), 1 µL cDNA, 0.5 µL each primer (10 µM), and 8 µL RNAse-free water. qPCR assays were performed using an ABI 7500 Fast Real-Time PCR system (Thermo Fisher Scientific). The PCR program began with a cycle at 95 °C for 2 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. For S. litura and S. littoralis, the qPCR mix contained 5 µL LightCycler 480 SYBR Green I Master (Roche, Basel, Switzerland), 4 µL diluted cDNA, and 0.5 µL of each primer. qPCR reactions were performed on a CFX384 Touch Real-Time PCR system (BioRad, Hercules, CA, USA) and the PCR program began with a cycle at 95 °C for 13.5 min, followed by 50 cycles at 95 °C for 10 s, 60 °C for 15 s, and 72 °C for 15 s. Dissociation curves of the amplified products were performed by gradual heating from 55 °C to 95 °C at 0.5 °C. s−1. The fivefold dilution series were used to construct relative standard curves to determine the PCR efficiencies used for quantification. SexiATPs, SfruActin, SlitRPL13, and SlituRPL13 were used as reference genes. Mean normalized expression of target genes compared to the reference gene was calculated with Q-Gene (52) using the following formula (NE: normalized expression; E: primer efficiency; CT: cycle threshold):
Functional Analysis in Drosophila Neurons.
The generation of the Drosophila line expressing SlitOr5 in at1 OSNs was previously described (17). The SfruOr5 full-length open-reading frame (ORF) was amplified from S. frugiperda male antennal cDNA, cloned into a pCRII-TOPO™ vector (Thermo Fisher Scientific), and subcloned into the pUAST.attB vector by BMGIF (CNRS, Gif-Sur-Yvette, France). The SexiOr5, SlituOr5, SlituOr75, SlitOr75, AncOr5, AncOr75, and AncOr5_75 full-length ORFs were synthesized in vitro and subcloned into the pUAST.attB vector by Synbio Technologies (Monmouth Junction, NJ, USA). For SexiOr5 and AncOr sequences, codons were optimized for expression in Drosophila. All transformant UAS-Or lines were generated by BestGene Inc. (Chino Hills, CA, USA), by injecting the pUAST.attB-Or plasmids (Endofree prepared, Qiagen, Hilden, Germany) into fly embryos with the genotype y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP}ZH-51C (53). The UAS-Or balanced lines were then crossed to the Or67dGAL4[2] line (23) to obtain double homozygous flies expressing the Or transgene in at1 OSNs instead of Or67d. Flies were reared on standard cornmeal–yeast–agar medium and kept in a climate- and light-controlled environment (25 °C, 12 h:12 h light:dark cycle).
A screening of 26 pheromone compounds (SI Appendix, Table S2) was performed by using single-sensillum extracellular recordings on at1 OSNs as previously described (54). The pheromone compounds were either synthesized in the lab or purchased from Sigma-Aldrich (St Louis, MO, USA) and Pherobank (Wijk bij Duurstede, The Netherlands) and were diluted in hexane (Carlo Erba Reagents, Val de Reuil, France). at1 OSNs were stimulated during 500 ms, using stimulus cartridges made of Pasteur pipettes containing 10 µg pheromone dropped onto a filter paper. Dose–response analyses were performed using the same methods, with doses ranging from 10 ng to 100 µg pheromone in the stimulus cartridge. Data were analyzed using GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, USA). Odorants were considered as active if the response they elicited was statistically different from the response elicited by the solvent alone (one-way ANOVA followed by a Dunnett’s post hoc test).
Functional Characterization in Xenopus Oocytes.
ORFs of SexiOr5, SfruOr5, SlituOr5, AncOr5, AncOr75, AncOr5_75, and SlitOrco (which was synthesized in vitro) were subcloned into the pCS2+ vector by Synbio Technologies. The template plasmids were fully linearized with NotI, and capped cRNAs were transcribed using SP6 RNA polymerase with the mMESSAGE mMACHINE SP6 transcription kit (Thermo Fisher Scientific). Purified cRNAs were resuspended in nuclease-free water at a 2 mg/mL concentration and stored at –80 °C until use. Stock solutions of the pheromone compounds tested were prepared by diluting each compound to 0.1 M in dimethyl sulfoxide (DMSO) and stored at −20 °C. Before each experiment, the stock solution was diluted to working concentration in 1× Ringer’s buffer (96 mM NaCl, 2 mM KCl, 5 mM MgCl2, 0.8 mM CaCl2, 5 mM HEPES, pH 7.6).
The functional characterization of receptors was performed by heterologous expression in Xenopus oocytes combined with two-electrode voltage-clamp. Inward currents in oocytes expressing the receptors SexiOR5, SfruOR5, and SlituOR5 were measured in response to a panel of 13 pheromone compounds (10–4 M) as described in ref. 55. Experiments on ancestral receptors and related mutant receptors (see above) were performed with a panel of six pheromone compounds as described in ref. 56. Ringer’s buffer containing DMSO was used as a negative control. Screening experiments were performed with compounds diluted at 10–5 M, and ascending pheromone concentrations from 10–8 to 10–4 M were used for dose–response experiments. All data were analyzed using GraphPad Prism 8. To compare responses between the different ancestors and mutants, we performed a principal component analysis to extract the main principal components that explain their variability in tuning using the FactoMineR package in R (57). Taken together, PC1 (84.94%) and PC2 (8.59%) explain more than 93% of the total variability and are sufficient to discriminate tunings of the different receptors. No clear relationship was visible between the principal components and the tuning breadth.
Reconstruction of Ancestral OR Sequences.
To reconstruct putative ancestral sequences of OR5 and OR75, a multiple sequence alignment was first performed using MAFFT (58) with homologous amino acid sequences from Noctuidae species, including S. littoralis, S. litura, S. exigua, S. frugiperda, Helicoverpa armigera, H. zea, Athetis dissimilis, and A. lepigone. Second, a phylogenetic tree was generated using PhyML (50) based on the resulting alignment. The ancestral reconstruction itself was done using the tool codeml of the package PAML (59) using both the topology of the phylogenetic tree obtained previously and the multiple sequence alignment. Briefly, codeml uses a maximum likelihood and an Empirical Bayes approach to estimate the likelihood of an ancestral sequence given the actual data, a tree topology, and a model of evolution. Using this method, the robustness of the ancestral reconstruction is evaluated for each amino acid position using a posterior probability. Due to the addition of distantly related sequences for the ancestral reconstruction, the sequences obtained for our nodes of interest were 398 amino acid long compared to the 397 amino acid sequence of the actual receptors. The additional leucine added by the reconstruction was then removed from the ancestral sequences. Values of posterior probabilities are ranging from 0.228 to 1, with more than 69% of sites showing posterior probabilities above 0.995 for AncOR5_75, and more than 92 and 93% of sites for AncOR5 and AncOR75, respectively (SI Appendix, Fig. S4).
Protein Modeling and Ligand-Binding Predictions.
The AlphaFold2 algorithm (60) was used to model the 3D structures of both current and ancestral ORs, through the Institut Français de Bioinformatique Core Cluster (ANR-11-INBS-0013). Best-ranked structures were then compared between orthologs using the MatchMaker tool of ChimeraX (61) and the rmsd computed using the same tool (61). The coordinates of putative binding pockets were identified with DeepSite (62). Webina (63) was used to dock known ligands to modeled structures using coordinates obtained from DeepSite, leading to nine different poses for each ligand. Residues located at <5 Å from all the nine poses of the ligand were considered as putatively interacting with it.
Site-Directed Mutagenesis.
Individual mutations were introduced in the pCS2+/AncOR5_75 plasmid using the Q5 Site-directed Mutagenesis kit (New England Biolabs, Ipswich, MA, USA) with primers designed using NEBaseChanger (https://nebasechanger.neb.com/, SI Appendix, Table S3). Double and triple mutation combinations were selected based on the 3D structure analysis as residues that are in close proximity to each other. Plasmids were checked for successful mutations by Sanger sequencing (MicroSynth AG, Balgach, Switzerland) and then mini-prepped using QIAprep Spin Miniprep kit (Qiagen). For the AncOR5_75 receptor carrying eight mutations (AncOR5_75_mut8x), the ORF was synthesized in vitro by Synbio Technologies and subcloned into the pCS2+ vector.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work has been funded by the French National Research Agency (ANR-16-CE02-0003, ANR-16-CE21-0002, ANR-22-CE13-0013, ANR-20-CE20-0003, 20-PCPA-0007 grants), the National Natural Science Foundation of China (32130089 and 31725023), a PRC NSFC-CNRS 2019-2021 grant, a CAAS Opening fund SKLOF202002, and the French Embassy in China (COMIX 2019-2020). We thank the Chinese Scholarship Council (CSC) for financial support of Z.L. PhD study (CSC Grant No.202003250076). Research was conducted as part of the CAAS-INRAE Associated International Laboratory in Plant Protection.
Author contributions
Y.L., G.W., E.J.-J., N.M., and C.M. designed research; Z.L., R.C., L.B.-H., S.Z., D.S., P.L., D.D.-M., M.-C.F., N.M., and C.M. performed research; N.M. and C.M. contributed new reagents/analytic tools; Z.L., R.C., L.B.-H., S.Z., D.D.-M., N.M., and C.M. analyzed data; and Z.L., N.M., and C.M. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. J.B. is a guest editor invited by the Editorial Board.
Contributor Information
Nicolas Montagné, Email: nicolas.montagne@sorbonne-universite.fr.
Camille Meslin, Email: camille.meslin@inrae.fr.
Data, Materials, and Software Availability
Data/codes have been deposited in Recherche Data Gouv (https://doi.org/10.57745/VB73LJ) (64). All other study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Allison J. D., Carde R. T., Pheromone Communication in Moths: Evolution, Behavior, and Application (University of California Press, 2016). 10.1525/9780520964433. Accessed 14 December 2022. [DOI] [Google Scholar]
- 2.De Pasqual C., Groot A. T., Mappes J., Burdfield-Steel E., Evolutionary importance of intraspecific variation in sex pheromones. Trends Ecol. Evol. 36, 848–859 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Jurenka R. A., “2 - lepidoptera: Female sex pheromone biosynthesis and its hormonal regulation” in Insect Pheromone Biochem. Mol. Biol., Blomquist G. J., Vogt R. G., Eds. (Academic Press, ed. 2, 2021), pp. 13–88. [Google Scholar]
- 4.Benton R., Sachse S., Michnick S. W., Vosshall L. B., Atypical membrane topology and heteromeric function of drosophila odorant receptors in vivo. PLoS Biol. 4, e20 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wicher D., et al. , Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Sato K., et al. , Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Zhang D.-D., Löfstedt C., Moth pheromone receptors: Gene sequences, function, and evolution. Front. Ecol. Evol. 3, 105 (2015). [Google Scholar]
- 8.Fleischer J., Krieger J., Insect pheromone receptors – key elements in sensing intraspecific chemical signals. Front. Cell. Neurosci. 12, 425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groot A. T., Dekker T., Heckel D. G., The genetic basis of pheromone evolution in moths. Annu. Rev. Entomol. 61, 99–117 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Nei M., Rooney A. P., Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39, 121–152 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Gracia A., et al. , Molecular evolution of the major chemosensory gene families in insects. Heredity 103, 208–216 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Montagné N., Wanner K., Jacquin-Joly E., “15 - Olfactory genomics within the lepidoptera” in Insect Pheromone Biochemistry and Molecular Biology, Blomquist G. J., Vogt R. G., Eds. (Academic Press, ed. 2, 2021), pp. 469–505. [Google Scholar]
- 13.Leary G. P., et al. , Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U. S. A. 109, 14006–14081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang K., Huang L.-Q., Ning C., Wang C.-Z., Two single-point mutations shift the ligand selectivity of a pheromone receptor between two closely related moth species. ELife 6, e29100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao S., Liu Y., Wang B., Wang G., A single point mutation causes one-way alteration of pheromone receptor function in two heliothis species. iScience 24, 102981 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Sayed A. M., The Pherobase: Database of pheromones and semiochemicals. www.pherobase.com. Accessed 29 November 2022.
- 17.Bastin-Héline L., et al. , A novel lineage of candidate pheromone receptors for sex communication in moths. ELife 8, 1–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quero C., Lucas P., Renou M., Guerrero A., Behavioral responses of spodoptera littoralis males to sex pheromone components and virgin females in wind tunnel. J. Chem. Ecol. 22, 1087–1102 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Meslin C., et al. , Spodoptera littoralis genome mining brings insights on the dynamic of expansion of gustatory receptors in polyphagous noctuidae. G3 (Bethesda) 12, jkac131 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouin A., et al. , Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 7, 11816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng T., et al. , Genomic adaptation to polyphagy and insecticides in a major east asian noctuid pest. Nat. Ecol. Evol. 1, 1747–1756 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Kergoat G. J., et al. , A novel reference dated phylogeny for the genus spodoptera guenée (Lepidoptera: Noctuidae: Noctuinae): New insights into the evolution of a pest-rich genus. Mol. Phylogenet. Evol. 161, 107161 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Kurtovic A., Widmer A., Dickson B. J., A single class of olfactory neurons mediates behavioural responses to a drosophila sex pheromone. Nature 446, 542–546 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Thornton J. W., Resurrecting ancient genes: Experimental analysis of extinct molecules. Nat. Rev. Genet. 5, 366–375 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Thornton J. W., Bridgham J. T., “Using ancestral gene resurrection to unravel the evolution of protein function” in Ancestral Sequence Reconstruction, Liberles D. A., Ed. (Oxford University Press, 2007). [Google Scholar]
- 26.Gu X., Zheng Y., Huang Y., Xu D., “Using ancestral sequence inference to determine the trend of functional divergence after gene duplication” in Ancestral Sequence Reconstruction (2007), pp. 117–127. [Google Scholar]
- 27.Yuvaraj J. K., et al. , Sex pheromone receptors of the light brown apple moth, epiphyas postvittana, support a second major pheromone receptor clade within the lepidoptera. Insect Biochem. Mol. Biol. 141, 103708 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Shen S., et al. , Evolution of sex pheromone receptors in dendrolimus punctatus walker (lepidoptera: Lasiocampidae) is divergent from other moth species. Insect Biochem. Mol. Biol. 122, 103375 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Kergoat G. J., et al. , Disentangling dispersal, vicariance and adaptive radiation patterns: A case study using armyworms in the pest genus spodoptera (Lepidoptera: Noctuidae). Mol. Phylogenet. Evol. 65, 855–870 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Campion D. G., et al. , Modification of the attractiveness of the primary pheromone component of the egyptian cotton leafworm, spodoptera littoralis (Boisduval) (Lepidoptera: Noctuidae), by secondary pheromone components and related chemicals. Bull. Entomol. Res. 70, 417–434 (1980). [Google Scholar]
- 31.Yan Q., et al. , Two sympatric spodoptera species could mutually recognize sex pheromone components for behavioral isolation. Front. Physiol. 10, 1256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C., et al. , Optimization of a pheromone lure by analyzing the peripheral coding of sex pheromones of spodoptera frugiperda in china. Pest Manag. Sci. 78, 2995–3004 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Poivet E., et al. , The use of the sex pheromone as an evolutionary solution to food source selection in caterpillars. Nat. Commun. 3, 1047 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Guo P.-P., et al. , The genetic basis of gene expression divergence in antennae of two closely related moth species, helicoverpa armigera and helicoverpa assulta. Int. J. Mol. Sci. 23, 10050 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phelan L., Evolution of sex pheromones and the role of asymmetric tracking. Evol. Sex Pheromones Role Asymmetric Track, 265–314 (1992). [Google Scholar]
- 36.Löfstedt C., Butlin R. K., Guilford T., Krebs J. R., Moth pheromone genetics and evolution. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 340, 167–177 (1997). [Google Scholar]
- 37.Pellegrino M., Steinbach N., Stensmyr M. C., Hansson B. S., Vosshall L. B., A natural polymorphism alters odour and DEET sensitivity in an insect odorant receptor. Nature 478, 511–514 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman S., Luetje C. W., Mutant cycle analysis identifies a ligand interaction site in an odorant receptor of the malaria vector anopheles gambiae. J. Biol. Chem. 292, 18916–18923 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes D. T., Wang G., Zwiebel L. J., Luetje C. W., A determinant of odorant specificity is located at the extracellular loop 2-transmembrane domain 4 interface of an anopheles gambiae odorant receptor subunit. Chem. Senses 39, 761–769 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuvaraj J. K., et al. , Putative ligand binding sites of two functionally characterized bark beetle odorant receptors. BMC Biol. 19, 1–21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franco F. P., Xu P., Harris B. J., Yarov-Yarovoy V., Leal W. S., Single amino acid residue mediates reciprocal specificity in two mosquito odorant receptors. Elife 11, e82922 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prieto-Godino L. L., Schmidt H. R., Benton R., Molecular reconstruction of recurrent evolutionary switching in olfactory receptor specificity. ELife 10, e69732 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schupfner M., Straub K., Busch F., Merkl R., Sterner R., Analysis of allosteric communication in a multienzyme complex by ancestral sequence reconstruction. Proc. Natl. Acad. Sci. U.S.A. 117, 346–354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voordeckers K., et al. , Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 10, e1001446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee C., Su B.-H., Tseng Y. J., Comparative studies of alphafold, RoseTTAFold and Modeller: A case study involving the use of G-protein-coupled receptors. Brief. Bioinform. 23, bbac308 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slater G., et al. , Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6, 31 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afgan E., et al. , The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu L., Niu B., Zhu Z., Wu S., Li W., CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guindon S., et al. , New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Anisimova M., Gascuel O., Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Simon P., Q-Gene: Processing quantitative real-time RT-PCR data. Bioinforma. Oxf. Engl. 19, 1439–1440 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., An optimized transgenesis system for drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Fouchier A., et al. , Evolution of two receptors detecting the same pheromone compound in crop pest moths of the genus spodoptera. Front. Ecol. Evol. 3, 1–11 (2015). [Google Scholar]
- 55.Cao S., Liu Y., Wang G., Protocol to identify ligands of odorant receptors using two-electrode voltage clamp combined with the xenopus oocytes heterologous expression system. STAR Protoc. 3, 101249 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vandroux P., et al. , Activation of pheromone-sensitive olfactory neurons by plant volatiles in the moth agrotis ipsilon does not occur at the level of the pheromone receptor protein. Front. Ecol. Evol. 10, 1143 (2022). [Google Scholar]
- 57.Lê S., Josse J., Husson F., FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 25, 7535 (2008). [Google Scholar]
- 58.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen E. F., et al. , UCSF chimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. Publ. Protein Soc. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiménez J., Doerr S., Martínez-Rosell G., Rose A. S., De Fabritiis G., DeepSite: Protein-binding site predictor using 3D-convolutional neural networks. Bioinformatics 33, 3036–3042 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Kochnev Y., Hellemann E., Cassidy K. C., Durrant J. D., Webina: An open-source library and web app that runs AutoDock Vina entirely in the web browser. Bioinformatics 36, 4513–4515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meslin C., Data for “A tale of two copies: evolutionary trajectories of moth pheromone receptors.” Recherche Data Gouv. 10.57745/VB73LJ. Deposited 25 April 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Data/codes have been deposited in Recherche Data Gouv (https://doi.org/10.57745/VB73LJ) (64). All other study data are included in the article and/or SI Appendix.


