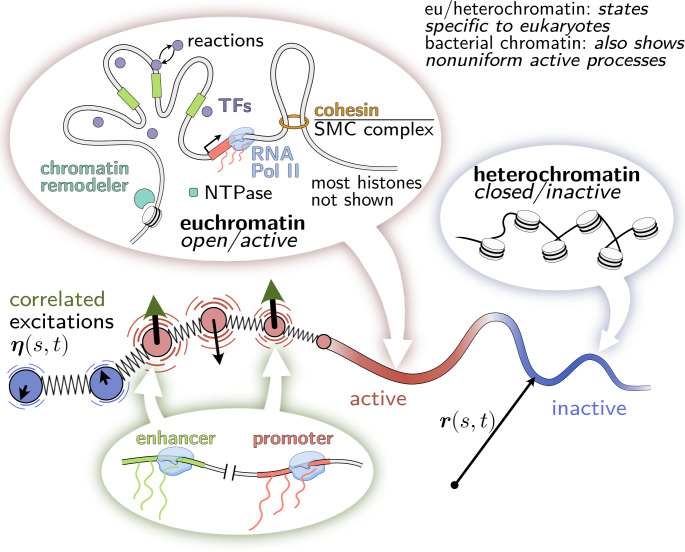
Fig. 1.
Model of the genome as an active heteropolymer. The genome acts as a scaffold for a myriad of active processes. Motor proteins, such as RNA polymerase II and loop extruding factors, consume chemical energy to exert forces on DNA (24). These nucleoside triphosphatases (NTPases) hydrolyze nucleoside triphosphates (NTP) such as ATP, thereby switching to an NDP-bound state, and restore their NTP-bound conformation by nucleotide exchange (28). For example, specialized proteins actively modify DNA-bound histones and transcription factors via nonequilibrium reactions (23, 29, 30). In the context of eukaryotes, active processes are enriched in transcriptionally active euchromatin compared to inactive heterochromatin. (For visual clarity in the figure, histones in euchromatin and active processes in heterochromatin are omitted.) In our coarse-grained model, we describe active processes by random forces that have larger magnitude and thus drive faster diffusion in active regions than in inactive regions. We also stipulate that the active forces at different loci are not necessarily independent. We hypothesize that correlated excitations could arise, for example, from the coordinated activation of an enhancer and its cognate promoter. Alternatively, correlated excitations could also arise from fluctuating concentration gradients of molecules that show sequence-specific interactions with chromatin.