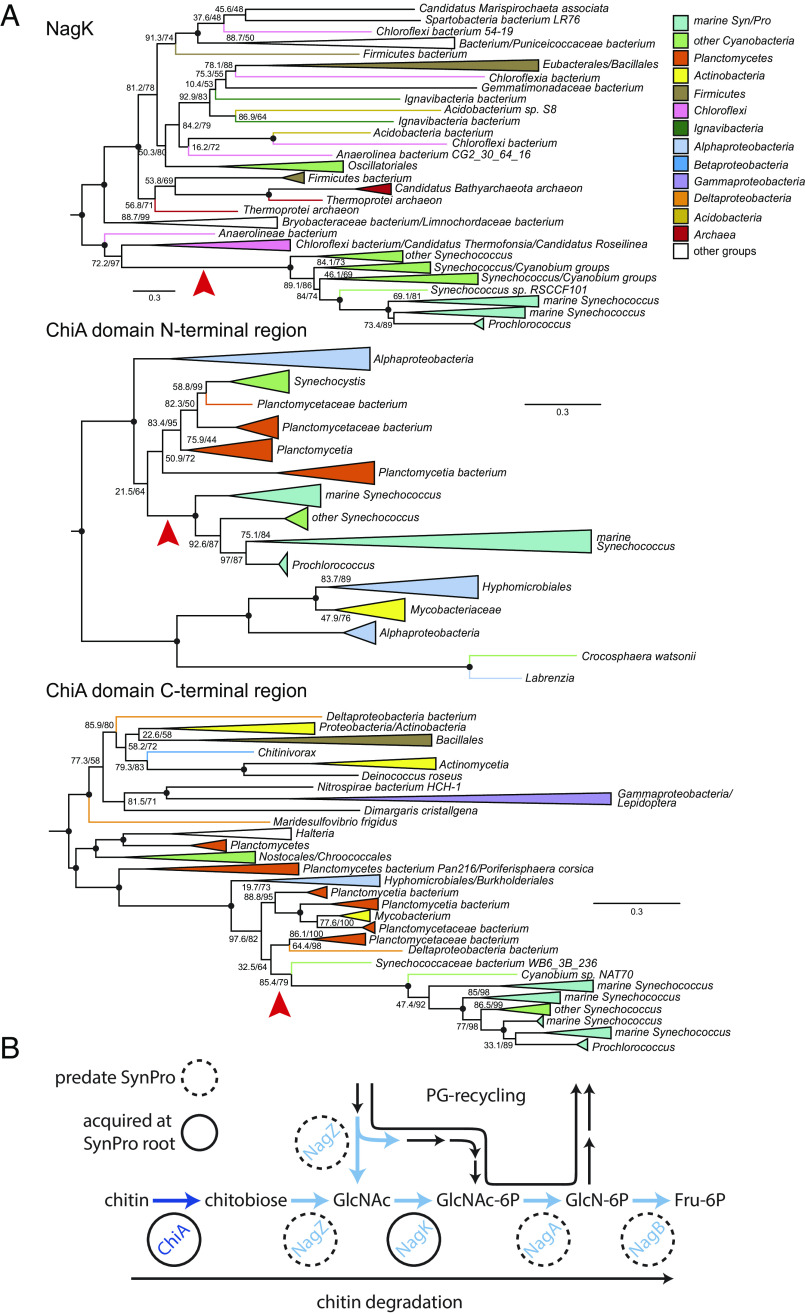
Fig. 6.
Phylogenies of ChiA and NagK genes in marine picocyanobacteria. (A) Red arrows indicate inferred HGTs into SynPro ancestor (stem) lineages. Branch lengths are indicated by included scale bars (average substitutions/site). High-level taxonomic identities are indicated by color coding, with specific represented groups labeled for terminal and collapsed groups. In each case, trees are rooted at the branch closest to the midpoint that preserves the monophyly of Synechococcus groups including SynPro. Branch supports (approximate likelihood ratio test/bootstrap value) are included for bipartitions with less than 90% support for either metric. Black circles at the internal node represent branches with both supports >90%. Phylogenies of NagZ, NagA, NagB, UgpA, and UgpE are found in SI Appendix, Fig. S8. (B) Pathways for chitin utilization and peptidoglycan metabolic recycling. Genes exclusive to chitin utilization are shown in dark blue, and genes shared between the two pathways are shown in blue. Genes highlighted with dashed circles predate evolution of marine picocyanobacteria, while genes highlighted with solid circles were acquired along the stem leading to their crown group.