Highlights
-
•
CRISPR/Cas9 system can improve the efficiency of HSV-1 genome editing.
-
•
CRISPR/Cas9 system successfully mediates the modification of therapeutic gene armed oncolytic herpes simplex virus (O-HSV1211).
-
•
IL12 and CXCL11 armed O-HSV1211 can significantly enhance the therapeutic outcome in Colorectal cancer.
-
•
The antitumor effect of IL12 and CXCL11 in O-HSV1211 is dependent on IFN-γ.
Keywords: CRISPR/Cas9 system, Herpes simplex virus-1, Genome editing, Oncolytic virus, Therapeutic gene, Colorectal cancer
Abstract
Oncolytic viruses are an emerging cancer treatment modality with promising results in clinical trials. The new generation of oncolytic viruses are genetically modified to enhance virus selectivity for tumor cells and allow local expression of therapeutic genes in tumors. The traditional technique for viral genome engineering based on homologous recombination using a bacterial artificial chromosome (BAC) system is laborious and time-consuming. With the advent of the CRISPR/Cas9 system, the efficiency of gene editing in human cells and other organisms has dramatically increased. In this report, we successfully applied the CRISPR/Cas9 technique to construct an HSV-based oncolytic virus, where the ICP34.5 coding region was replaced with the therapeutic genes murine interleukin 12 (IL12, p40-p35) and C-X-C motif chemokine ligand 11 (CXCL11), and ICP47 gene was deleted. The combination of IL12 and CXCL11 in oncolytic viruses showed considerable promise in colorectal cancer (CRC) treatment. Overall, our study describes genetic modification of the HSV-1 genome using the CRISPR/Cas9 system and provides evidence from principle studies for engineering of the HSV genome to express foreign genes.
1. Introduction
Herpes simplex virus type 1 (HSV-1) is a highly prevalent human pathogen that infects or is carried by nearly 90% of the population (Whitley and Roizman, 2001). HSV-1 is composed of a 152-kb linear double-stranded DNA that allocates approximately 84 open reading frames arranged for contiguous transcription (Martinez et al., 1996). The HSV-1 genome can accommodate almost 40 kb of foreign genes, such as intergenic regions and nonessential gene regions. Several HSV-1 genes play important roles against host immune responses. For instance, ICP34.5, a neurovirulent factor, targets stimulator of interferon genes (STING), leading to downregulation of interferon regulatory factor 3 (IRF3) and the production of type I interferon (IFN-α/β) (Pan et al., 2019). ICP34.5 also counteracts dsRNA-dependent protein kinase (PKR) and maintains the active state of the α subunit of translation initiation factor eIF2 (eIF2-α) for viral protein synthesis (Li et al., 2011; Tallóczy et al., 2014). Previous studies have demonstrated that ICP34.5-null mutant viruses, such as R3616, fail to resist interferon-α/β and thus attenuate virulence (Cheng et al., 2002). In addition, ICP34.5 downregulates CD86 expression on dendritic cells (DCs) in infectious animal models, indirectly affecting the adaptive immunity of the host (Jin et al., 2011). In addition, ICP47 inhibits MHC-I antigen presentation by binding transporter-associated protein (TAP) and hinders the adaptive immune response of the host (Matschulla et al., 2017; Raafat et al., 2012). Understanding the mechanism of viral genes is important for modification of HSV-1 as a vehicle for therapeutic applications.
The first-generation HSV-1-based oncolytic virus, G207 (Mineta et al., 1995), with deleted ICP34.5 and UL39 genes, demonstrated antitumor activity in the treatment of malignant glioma (Post1 et al., 2004). Furthermore, deletion of the ICP47 gene in G207 permits immediate early expression of US11, which inhibits PKR and favors replication of the ICP34.5-deficient G207 virus (Apetoh et al., 2010; Fukuhara et al., 2016; Poppers et al., 2000; Todo et al., 2001). The development of HSV-1-based oncolytic viruses attracted extensive attention with the FDA approval of the HSV-1-based oncolytic virus T-Vec (talimogene laherparepvec) for the treatment of metastatic melanoma in 2015. T-Vec not only deletes the above neurovirulence factors but also carries the coding sequence of granulocyte macrophage colony-stimulating factor (GM-CSF) (Bommareddy et al., 2016; Harrington et al., 2010; Liu et al., 2003). To explore more options to modify the tumor microenvironment, a new generation of oncolytic HSV viruses expressing a range of immunostimulatory modulators are to be prepared and tested. Therefore, a convenient and efficient approach to engineer oncolytic viral vehicles is in high demand.
Previously, HSV-1 viruses were engineered using a bacterial artificial chromosome (BAC) system. However, construction of a BAC vector for recombination, such as transformant growth and selection, is time-consuming. To accommodate the BAC vector, the genes were truncated, which may cause replication deficiency (Agarwalla and Aghi, 2012; Gierasch et al., 2006). The CRISPR/Cas9 system has a substantial advantage in site-specific gene editing and has been successfully applied to modify the HSV-1 genome (Gao et al., 2014; Hsu et al., 2014, 2013; Russell et al., 2015). A double-strand DNA break (DSB) at a designated site was induced by gRNA and repaired via homology-directed repair (HDR). Although this approach has been successfully established for foreign gene insertion into the HSV-1 genome, it is still limited to fluorescence selection markers (Lin et al., 2016). However, the genomic modification method for HSV-based oncolytic viruses and the efficiency of the CRISPR/Cas9 system still need to be explored and optimized.
Here, we describe the construction of HSV-based oncolytic viruses by CRISPR/Cas9-directed homologous recombination. We also inserted an IRES sequence to coexpress the therapeutic genes IL12 (p40-p35) and CXCL11 to facilitate the treatment of colon tumors by genetically modified HSV-1.
2. Materials and methods
2.1. Cell lines and viruses
HEK293FT cells, human skin fibroblast (HSF) cells, SW620 cells, MC38 cells and Vero cells were originally from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, 12800-017) with 10% FBS at 37 °C and 5% CO2. The original HSV-1 (F strain) virus was from Roizman's lab (He et al., 1997), and all viruses were grown and titrated on Vero cells.
Mouse CD8+ and CD4+ T cells were purified from the peripheral lymph nodes and spleens by CD8a (Ly-2) MicroBeads (Miltenyi, 130-117-044) and CD4 (L3T4) MicroBeads (Miltenyi, 130-117-043). T cells were cultured in plates coated with 10 µg/ml anti-CD3 (BioXCell, 145-2C11) and 5 µg/ml anti-CD28 (BioXCell, 37.51). T cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 1% sodium pyruvate, 1% penicillin‒streptomycin, and 50 μM 2-mercaptoethanol.
Other viruses were generated by our lab as follows: the HSV-GFP virus contains a GFP cassette in the intergenic region of the UL26 and UL27 genes; D-34.5 contains a GFP cassette that replaces the ICP34.5 gene in the HSV-1 genome; O-HSV1 was obtained by deleting the ICP47 gene in the D-34.5 genome; and O-HSV11, O-HSV12 and O-HSV1211 was obtained by replacing the GFP cassette with the CXCL11, IL12 and IL12-IRES-CXCL11 cassette in O-HSV1. These viruses were constructed by the CRISPR/Cas9 system following several rounds of plaque purification under a fluorescence microscope. Transfections were carried out on HEK293FT cells with Lipofectamine 2000 (Invitrogen, USA). HEK293FT cells were cultured at a density of 5×105 cells/well in 6-well plates for 24 h and transfected with plasmids (Donor plasmid: sgRNA plasmid=1:1). After 24 h of incubation (37 °C, 5% CO2), the cells were infected with HSV-1 or mutant HSV-1 at a multiplicity of infection (MOI) of 1. After 48 h of infection, all cell-associated viruses were harvested with 9% milk and by three cycles of freezing and thawing to lyse the cells. Virus samples were serially diluted and used to infect Vero cells. At 48 h postinfection (hpi), single virus plaques were selected under a fluorescence microscope.
2.2. Antibodies and reagents
The antibodies and regents used in this study are listed as follows: ICP34.5 antibody provided by Dr. B. He (University of Illinois at Chicago, USA). IL12 antibody (Santa Cruz Biotechnology, sc-365389), CXCL11 antibody (ThermoFisher, PA5-47767), HRP-conjugated secondary antibodies (Sungene Biotech, catalog.no. LK2001 and LK2003), GDPDH antibody (Sungene Biotech, KM9002), IFN-γ antibody (BioXcell, BE0055), IFN-α (Sigma, SRP4596), and the Annexin V-FITC/PI Apoptosis Detection Kit (Solarbio, CA1020).
2.3. Plasmid construction
The targeting plasmid of the CRISPR/Cas9 system was constructed by annealing two complimentary oligonucleotides of sgRNA and ligating the dsDNA into the lentiCRISPR v2 plasmid by BsmbI.
For construction of the pICP34.5-HA2L-HA2R plasmid, the homology arms flanking the coding region of ICP34.5, HomologyArm2L (HA2L) and HomologyArm2R (HA2R), were amplified from HSV-1 DNA by PCR. Multiple clone sites with two complimentary oligonucleotides (oligo1 and oligo2) were annealed into the pCMV-HA vector between Sbf1 and EcoRI to obtain the pVector. HA2L and HA2R of ICP34.5 were sequentially cloned into pVector with SbfI/SpeI and PacI/EcoRI restriction sites to obtain pICP34.5-HA2L-HA2R. pCMV-eGFP was prepared in our laboratory by cloning the eGFP gene into the pCMV-HA vector at the SfiI/NotI restriction site. The GFP cassette (CMV-GFP-SV40PolyA) was amplified from pCMV-eGFP by PCR and inserted between HomologyArm2L and HomologyArm2R of pICP34.5-HA2L-HA2R with SpeI and PacI restriction sites to obtain the donor plasmid pICP34.5-HA2L-GFP-cassette-HA2R. All donor plasmids were constructed by the method described above, such as p2627-HA1L-GFP-cassette-HA1R, pICP47-HA3L-HA3R, pICP47-HA3L-RFP-HA3R, pICP34.5-HA2L-CXCL11-cassette-HA2R, pICP34.5-HA2L-IL12-cassette-HA2R and pICP34.5-HA2L-IL12-IRES-CXCL11-cassette-HA2R. The murine single-chain IL-12 (p40-p35) protein contains dual subunits involving p40 and p35 which were separated by an 18 amino-acid polypeptide linker (Lee et al., 2001). All sgRNA oligos and primer sequence information are listed in Supplemental table.
2.4. Titration of viruses
Confluent Vero cells were infected with tenfold serially diluted freeze‒thawed virus in 6-well plates. Plaque numbers were counted at 48 h postinfection, and viral titers were determined by multiplying the number of plaques by the dilution factor.
2.5. Cell proliferation assay
Cells were cultured on 96-well plates at a density of 5×104 cells per well. When cells were grown to 80% confluence, they were infected with the indicated viruses at an MOI = 1. At the indicated time points, the cells were incubated with Cell Counting Kit-8 (CCK8) (Solarbio, CA1210) for 30 min at 37 °C. The absorbance at a wavelength of 450 nm was determined with a microplate reader (Synergy 4; BioTek). The percentage of cell viability was calculated as follows: (OD value of infected cells-OD value of Black)/(OD value of uninfected cells-OD value of Black) × 100%.
2.6. Western blot analysis
Cell lysates were run on SDS polyacrylamide gel electrophoresis (SDS‒PAGE) gels and blotted onto polyvinylidene fluoride (PVDF) membranes. Western blotting was carried out using standard procedures. The quantitation of the protein bands was analyzed by Quantity One software (Bio-Rad).
2.7. RT‒PCR analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, 15596018). cDNA was synthesized using a HiScript II Q RT SuperMix for qPCR kit (Vazyme, R223-01) and by real-time PCR with a QuantStudio™ 7 Flex system. Primer information is listed in the Supplemental Table.
2.8. Preparation of tumor lymphocytes
Mice were inoculated with viable MC38 cells on Day 0 and i.t. treated with PBS or oncolytic viruses on Days 7, 10, and 13. Tumor-bearing mice were sacrificed on the indicated days, and single cells from tumor tissues were resuspended with 40% Percoll gradient medium and overlaid on 70% Percoll. The suspensions were centrifuged at 1260 g for 20 min (25 °C) without interruption. Cells in the interface between the 40% and 70% Percoll were used as lymphocytes. The frequencies of CD8 and CD4 T cells were determined with fluorescently conjugated antibodies and analyzed by flow cytometry.
To measure cytokine production, isolated lymphocytes were incubated at 37 °C for 3 h in 96-well flat-bottom plates in the presence of 50 ng/ml phorbol 12-myristate-13-acetate (Sigma‒Aldrich, P8139) and 500 ng/ml ionomycin (Sigma‒Aldrich, I0634) with 1 μg/ml brefeldin A (Sigma‒Aldrich, B6542). After surface staining, the Fixation/Permeabilization Solution Kit (BD, 554714) was used for cytokine detection. For transcription factor staining, isolated lymphocytes were stained with a surface marker, and nuclear factor staining was performed according to the manufacturer's protocol (eBioscience, 00-5523). Fluorescently conjugated antibodies against cell-surface, intracellular and nuclear antigens were used as follows: CD4-BV605 (Biolegend, 100547), CD8-AF700 (Biolegend, 100730), CD8-PB (Biolegend, 100728), IFN-γ-FITC (Biolegend, 505806), IL4-PE (Biolegend, 504103), IL9-APC (Biolegend, 514104) and Foxp3-AF647 (Biolegend, 126408). Flow cytometric analysis was performed on a CytoFlex (Beckman Coulter) using FlowJo software.
2.9. T cell coculture assay
SW620 and MC38 cells were cultured at a density of 5×105 cells/well for 48 h in 6-well plates, and mouse CD8+ or CD4+ T cells were cultured at a density of 1×106 cells/well for 24 h in 24-well plates precoated with anti-CD3 and anti-CD28 antibodies.
SW620 and MC38 cells were infected with O-HSV1 and O-HSV1211 at an MOI of 1, and then activated mouse CD8+ or CD4+ T cells (2×106 cells/well) were cocultured with infected SW620 or MC38 cells for 48 h. The cell numbers were counted under a microscope. For IFN-γ neutralization, the culture medium of CD8+ or CD4+ T cells was supplemented with anti-IFN-γ antibodies (BioXCell, clone XMG1.2).
2.10. Animal experiments
Animal experiments were carried out according to the Guide for the Care and Use of Laboratory Animals. Female C57BL/6J mice (6 weeks old) were purchased from Charles River (Beijing, China). For the establishment of syngeneic tumors, 1×105 MC38 cells in 100 µL DMEM were subcutaneously injected into the right flank of each mouse. When tumors reached 70–100 mm3 in size, mice were divided randomly into 3 or 4 groups (six mice per group). Viruses (1×107 PFUs per mouse) or PBS was injected into the tumors.
For IFN-γ blockade, mice were injected intraperitoneally (i.p.) with anti-IFN-γ antibodies (clone XMG1.2, BioXCell) or isotype control rat IgG1 (BE0088; BioXCell) antibodies on Days 7, 10, 13 and 16 post tumor implantation. The tumor volume (mm3) was measured by a Vernier caliper every 3 days and calculated as (length × width2)/2. When tumor volumes reached 2000 mm3, the mice were sacrificed.
2.11. Statistical analysis
All quantitative data are expressed as the mean ± SD of three independent experiments. Data were analyzed using Student's t test. Animal survival was presented using Kaplan‒Meier survival curves, and data for survival were analyzed by the log-rank test. A p value less than 0.05 (*) was considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
3. Results
3.1. CRISPR/Cas9 mediated the insertion of foreign genes in the intergenic regions of UL26 and UL27 of HSV-1
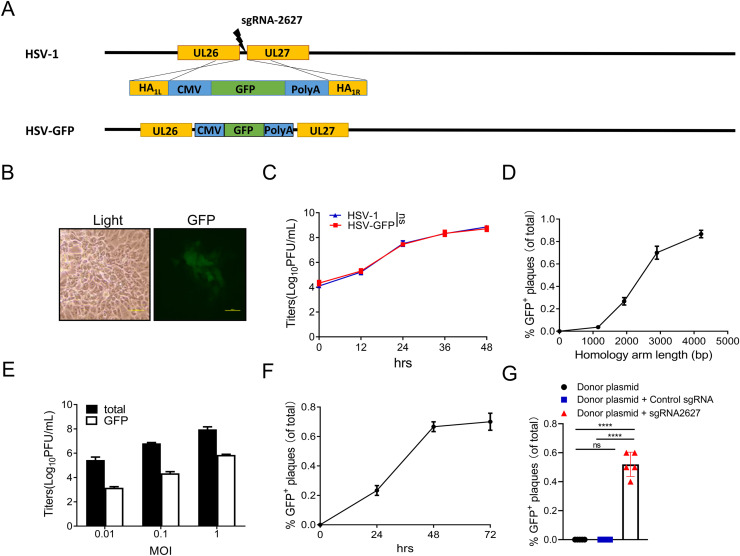
To develop the approach for CRISPR/Cas9-mediated HSV-1 genome editing, we first inserted a foreign gene in the intergenic region of UL26-UL27. sgRNA-2627 was designed to target and cleave dsDNA at the intergenic region of UL26-UL27. The GFP expression cassette was inserted into the UL26-UL27 intergenic regions of HSV-1 by homologous recombination of cleaved genome DNA with the donor plasmid, which included a 1471-bp left homology arm, GFP expression cassette and 1339-bp right homology arm (Fig. 1A). After virus genome editing and selection (Fig. 1B), we successfully obtained the recombinant virus HSV-GFP. The modified virus exhibited the same growth curve as the wild-type HSV-1, indicating that this insertion has no impact on viral replication (Fig. 1C).
Fig. 1.
Optimization of CRISPR/Cas9 -mediated modification of the HSV genome. A CRISPR/Cas9-mediated insertion of a foreign gene into the UL26–UL27 intergenic region. The intergenic region of UL26-UL27 was cleaved by Cas9, and the GFP cassette was inserted into the UL26-UL27 intergenic region by the homologous recombination of cleaved genome DNA with the donor plasmid. HA: homology Arm. B GFP-positive plaques were observed and selected under a fluorescence microscope (scale bar, 500 µm). C Viral replication of parental HSV-1 and HSV-GFP was measured with virus plaque titration and compared. D 293FT monolayers were transfected with donor plasmids containing different lengths of homology arm and sgRNA and infected with HSV-1 at an MOI of 1. Viruses were harvested at 48 h postinfection. E 293FT monolayers were transfected with donor plasmid and sgRNA-2627 and infected with HSV-1 at different MOIs. Virus was harvested at 48 h postinfection. F 293FT monolayers were transfected with donor plasmid and sgRNA-2627 and infected with HSV-1 at an MOI of 1. Viruses were harvested at different times postinfection. G 293FT monolayers were transfected with donor plasmid, donor plasmid plus control sgRNA or donor plasmid plus sgRNA2627 and infected with HSV-1 at an MOI of 1. Virus was harvested at 48 h postinfection. Virus plaque titration was performed on Vero cells. The numbers of total and GFP+ plaques were counted. P values were determined using Student's t test. ns, not significant, ****p < 0.0001.
Site-specific manipulation of the viral genome relies on the efficiency of homologous recombination between the virus genome and donor DNA. Inserting a GFP expression cassette in the intergenic region of UL26-UL27 was used as an example to explore the optimal conditions for gene editing of the HSV-1 genome. First, the different lengths of homology arms in the donor plasmid were tested. 293T cells were cotransfected with sgRNA-2627 and donor plasmid and then infected with HSV-1. The percentages of successfully modified viruses were calculated by infecting Vero cells with virus and counting plaques expressing green fluorescence protein under fluorescence microscopy. The frequency of successful gene editing was higher as the lengths of the homology arms increased (Fig. 1D). Second, we tested whether the recombination rate is affected by the multiplicity of infection (MOI) of HSV-1. After cotransfection with the donor plasmid with 1.5 kb homology arms and sgRNA-2627, 293T cells were infected with different amounts of HSV-1 (Fig. 1E). The results showed that the MOI of HSV-1 has a limited effect on the efficiency of gene editing of the HSV-1 genome. Third, increasing the culture time post HSV-1 infection improved the recombination efficiency, but a time longer than 48 hours post HSV-1 infection did not further increase recombination efficiency (Fig. 1F). However, without the assistance of the CRISPR/Cas9 system, the number of GFP+ plaques was significantly decreased only in the donor plasmid group, with a number 10,000 times lower than that in the donor plasmid plus sgRNA-2627 group (Fig. 1G). Our data indicated that the CRISPR/Cas9 system can be a powerful tool for HSV-1 genome editing. These optimized conditions were used for all subsequent experiments.
3.2. Engineering HSV-based oncolytic viruses by the CRISPR/Cas9 system
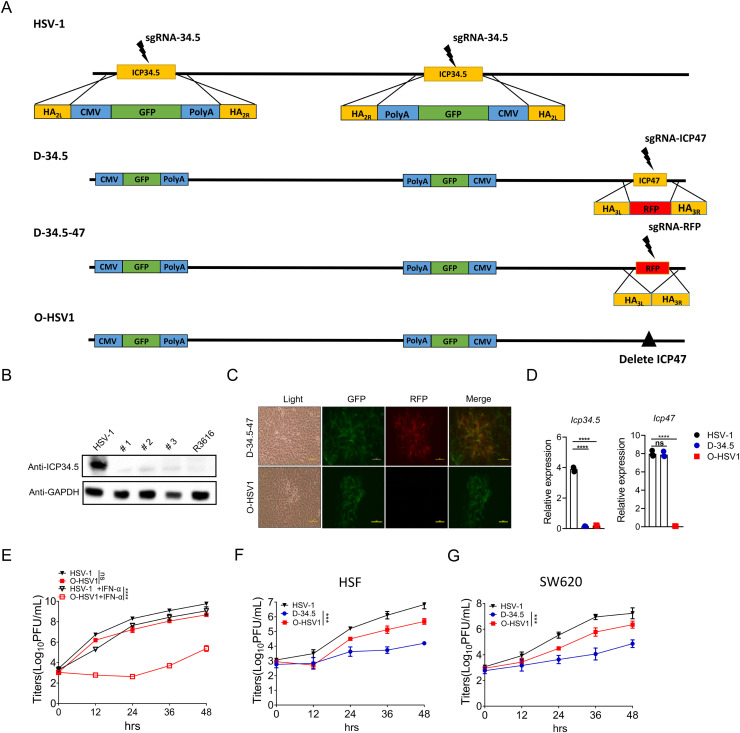
Elimination of virulent genes enables HSV-1 to replicate selectively in tumor cells to increase the safety prolife. Here, HSV-1 was used as a parental virus for the development of an HSV-based oncolytic virus by deleting two viral genes, ICP34.5 and ICP47. As shown in Fig. 2A, sgRNA-34.5 and donor plasmid pICP34.5-HA2L-GFP cassette-HA2R were designed to replace both copies of the ICP34.5 gene with the GFP cassette in the HSV-1 genome. The resulting GFP-expressing viruses, named D-34.5, were selected under a fluorescence microscope, and the deletion of ICP34.5 was examined by Western blot using antibodies as indicated (Fig. 2B). Wild-type HSV-1 was used as a positive control.
Fig. 2.
Engineering HSV-1 for an oncolytic virus by deleting the ICP34.5 and ICP47 genes. A Workflow of the gene editing process of the O-HSV1 oncolytic virus in this study. B 293FT monolayers were infected with HSV-1, O-HSV1 (#1/#2/#3) and R3616 at an MOI of 1. ICP34.5 protein expression was detected by Western blot. C Recombinant oncolytic virus O-HSV1 was obtained from two rounds of selection of RFP+ and RFP− plaques under a fluorescence microscope (scale bar at 500 µm). D Detection of the mRNA levels of ICP34.5 and ICP47 after virus infection in Hela cells by quantitative RT‒PCR. E Monolayers of Vero cells were untreated or pretreated with IFN-α before infection with HSV-1 and O-HSV1 virus. Viral titers were determined on Vero cells at different times postinfection (n=3). Human skin fibroblast (HSF) cells (F) and SW620 cells (G) were infected with different viruses, and virus replication was determined by a virus titration plaque assay (n=3). P values were determined using Student's t test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The ICP47 gene was subsequently deleted from D-34.5 in two steps. First, pICP47-HA3L-RFP-HA3R and sgRNA-47 were designed to replace the ICP47 gene of D-34.5 with red fluorescence protein (RFP) as a selection marker (Fig. 2A second row). The RFP-expressing plaques were selected under a fluorescence microscope and named D-34.5-47 (Fig. 2A third row). Second, pICP47-HA3L-HA3R and sgRNA-RFP were used to delete the RFP gene from D-34.5-47, and RFP-negative plaques were selected under a microscope (Fig. 2A bottom row and 2C). The proportion of recombinant viruses was approximately 40%–60% of the total number of RFP-negative viruses. The resulting oncolytic virus with deletion of both ICP34.5 and ICP47 was named O-HSV1 (clone #1/#2/#3) (Fig. 2D). All 3 clones of O-HSV1 were characterized by genotyping, sequencing, western blotting and virus titration assays (data not shown), which indicated no differences between the three clones; thus, clone 1 was further used for subsequent experiments. Intensive studies indicate that ICP34.5 is a virulent factor responsible for resistance against host innate immunity, such as type I interferon production and its antiviral signaling (Korn et al., 2009; Li et al., 2011). As shown in Fig. 2E, in the presence of IFN-α, O-HSV1 showed a defective replication capacity compared with that of wild-type HSV-1. Although compromised in growth compare with HSV-1, the oncolytic virus O-HSV1 with double deletion of the ICP34.5 and ICP47 genes replicated more efficiently than the ICP34.5-deleted virus D-34.5 (Fig. 2F and G).
3.3. O-HSV1 inhibited proliferation and induced cell death of human colon cancer cells in vitro
Colorectal cancer (CRC) is the third most commonly diagnosed malignant cancer worldwide and the second leading cause of death (Ganesh et al., 2019). Immunotherapy has achieved considerable progress in helping the immune system destroy cancer cells, such as immune checkpoint inhibitors (ICIs), CAR-T cells, tumor vaccines and immunostimulatory cytokines. However, the tumor microenvironment of CRC may form a closed barrier to inhibit the infiltration of immune cells and facilitate the initiation and progression of tumors. The established barrier of the TME compromises the treatment outcomes of immunotherapies (Zhang et al., 2020). Oncolytic viruses represent an alternative strategy to kill tumor cells by viral infection, which promotes the release of tumor-associated antigens and stimulates the immune response.
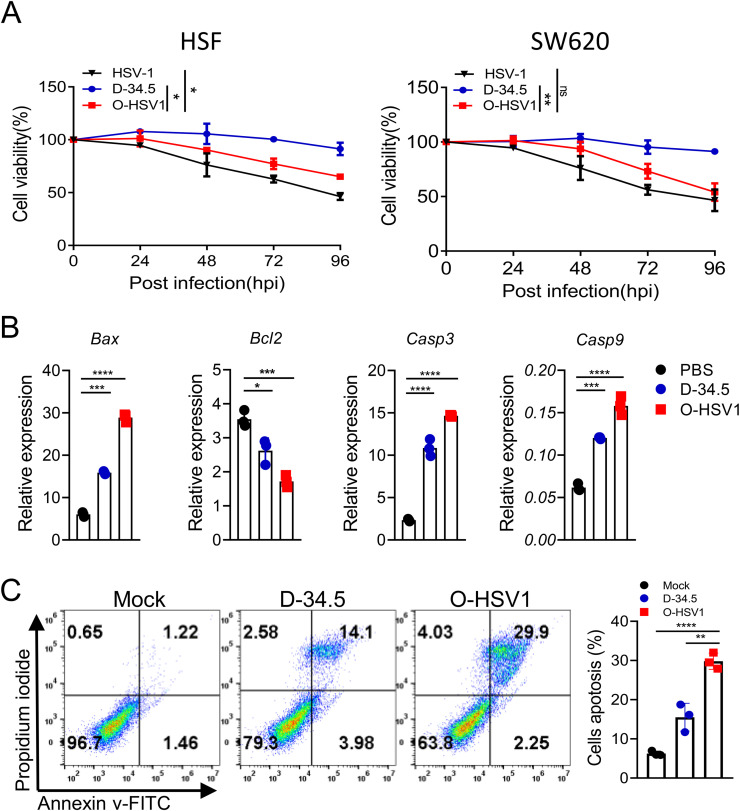
In vitro, O-HSV1 showed a high cytotoxicity in human CRC cell lines. To investigate the antitumor effect of O-HSV1, primary cultured SW620 or HSF cells were infected with HSV-1, D-34.5 and O-HSV1 at an MOI of 1, and cell viability was measured by CCK8 assay. As is shown in Fig. 3A, O-HSV1 infection resulted in an approximately 50% decrease in cell viability at 96 h. Interestingly, O-HSV1 showed defective replication efficiency in normal human fibroblast cells, whereas the inhibitory effect was comparable to that of HSV-1 in colon cancer cells. To explore whether apoptosis contributes to O-HSV1-induced cell death, tumor cells were subjected to quantitative RT‒PCR after infection with different viruses. As shown in Fig. 3B, the expression of apoptosis-associated genes (Bax, Casp3 and Casp9) was upregulated, while the negative regulator Bcl2 was downregulated. Furthermore, the apoptosis of SW620 cells induced by different viruses was detected by staining the apoptotic cell membrane with Annexin V-FITC. Thirty percent cell apoptosis was detected for O-HSV1 compared with 16% for D-34.5 (Fig. 3C), indicating that O-HSV1 exhibited an efficient oncolytic effect in colon cancer cells.
Fig. 3.
Oncolytic virus O-HSV1 inhibits proliferation and promotes apoptosis of SW620 cells. A Cell viability was measured by CCK8 assay after infection with different viruses at an MOI of 1. Uninfected cells were considered as 100% viable (n = 3). B Quantitative RT‒PCR analysis of Bax, Bcl2, Casp3 and Casp9 mRNA in SW620 cells infected with D-34.5 and O-HSV1 at an MOI of 1 for 48 h. C SW620 cell apoptosis induced by different virus was analyzed by Annexin V-FITC and PI staining. (n=3). ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (two-tailed Student's t test).
3.4. Construction of therapeutic gene-manipulated oncolytic virus O-HSV1211 by the CRISPR/Cas9 system
An adaptive immune response is critical for tumor obliteration. In particular, CD8+ T cells (cytotoxic T lymphocytes, CTLs) and T helper (Th) 1 cells, which are characterized by IFN-γ secretion, predominantly exhibit cytotoxicity against tumor cells. Evidence indicates that IL12 induced the production of IFN-γ from T cells not only acts on tumor cells to enhance the recognition by CD8+ and CD4+ T cells (Castro et al., 2018), but also promotes the cytotoxicity of CD8+ and CD4+ T cells (Bhat et al., 2017; Curtsinger et al., 2012; Takeda et al., 2017). Furthermore, infiltration of CTLs and Th1 cells into the tumor microenvironment (TME), which is largely dependent on the interaction of their chemokine receptors with chemokines, is of vital importance for the antitumor immune response. CXCL9, CXCL10 and CXCL11 via CXCR3 ligand have potent antitumor activity through the recruitment of CTLs and Th1 cells into the TME. CXCL11 is generally recognized as an important determinant of TME entry, and CXCL11-armed oncolytic poxvirus has exhibited value in antitumor therapy (Liu et al., 2016).
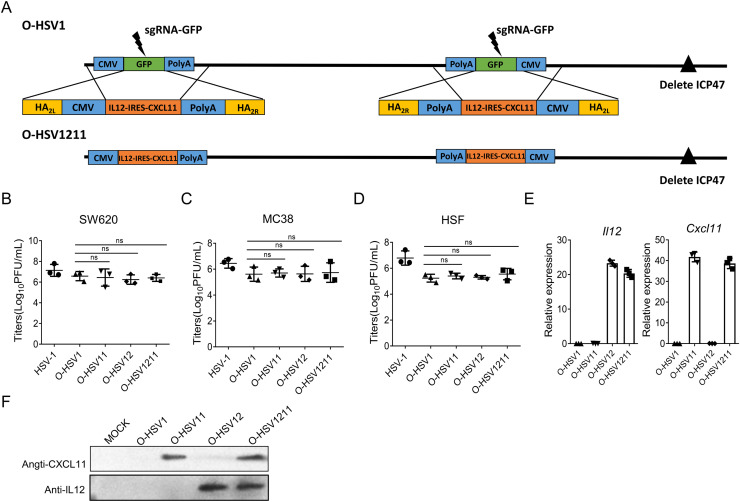
Based on the characteristics of IL12 and CXCL11, we hypothesize that the cooperation of IL12, CXCL11 and O-HSV1 may further improve oncolytic efficiency. To this end, we attempted to modify murine IL12 and CXCL11 expressing O-HSV1. The sgRNA-GFP was designed to target the GFP gene in O-HSV1. O-HSV1211 was generated by replacing GFP in O-HSV1 with IL12-IRES-CXCR11 using the donor plasmid pICP34.5-HA2L-IL12-IRES-CXCR11-cassette-HA2R (Fig. 4A). Accordingly, CXCL11 and IL12 armed oncolytic viruses, O-HSV11 and O-HSV12, were also generated by using this approach. GFP-negative plaques were isolated under a fluorescence microscope and different clones of O-HSV11(clone #1 #2), O-HSV12 (clone #1 #2 #3) and O-HSV1211 (clone #1 #2) were confirmed by genotyping and sequencing (data not shown). Recombinant viruses were used to infect SW620, MC38 (a colon cancer line of the C57BL/6 mice background) and HSF cells, and quantitative RT-PCR, western blot and virus titration assays were performed, which indicated that the expression of CXCL11, IL12 or IL12 and CXCR11 does not affect the viral replication compared with that of its parental virus (Fig. 4B–F). Clone 1 of O-HSV11, and O-HSV12 and O-HSV1211 was used for subsequent experiments. In this part, we present an evidence from a principle study to demonstrate that the introduction of foreign genes by the CRISPR/Cas9 system and improves the efficiency of viral genome manipulation, which may dramatically increase the efficiency of constructing diverse oncolytic viruses as potential cancer treatments.
Fig. 4.
Construction and characterization of the oncolytic virus O-HSV1211. A Schematic diagram of the construction of the oncolytic virus O-HSV1211. B–D SW620, MC38 and HSF cells were infected with HSV-1, O-HSV1, O-HSV11, O-HSV12 and O-HSV1211 at an MOI of 1, and titer were determined on Vero cells at 48 h postinfection. E and F Identification of IL12 and CXCL11 expression in O-HSV1, O-HSV11, O-HSV12 and O-HSV1211-infected MC38 cells. E Quantitative RT‒PCR analysis of Il12 and Cxcl11 mRNA in MC38 cells infected with oncolytic viruses at an MOI of 1 for 48 h. F IL12 and CXCL11 secretion in the medium was detected by western blotting. ns, not significant.
3.5. IL12 and CXCL11 expression promotes the antitumor capacity of oncolytic viruses
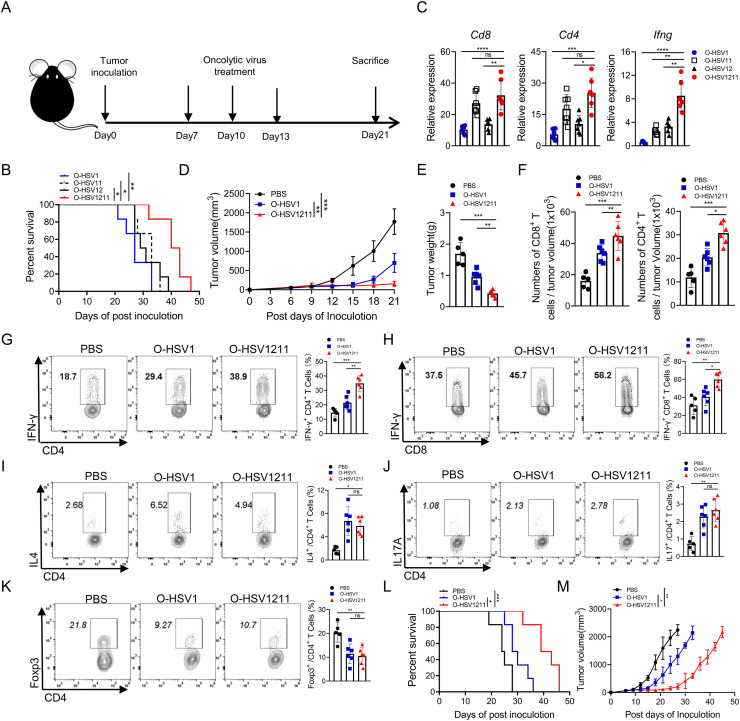
To examine the in vivo oncolytic effect of our modified oncolytic virus, an MC38 syngeneic tumor model was constructed. As shown in Fig. 5A, mice were treated with O-HSV1, O-HSV11, O-HSV12 and O-HSV1211 on Days 7, 10, and 13. The survival curve indicated that the median survival of O-HSV11 (armed CXCL11) and O-HSV12 (armed IL12)-treated group had a little increased than that in O-HSV1 treated-group (survived 4 or 5 more days), whereas the treatment of O-HSV1211 (armed IL12 and CXCL11) showed apparent advantage over O-HSV1(Fig. 5B). We further performed quantitative RT-PCR to analyze the expression of Cd8, Cd4 and Ifng in tumor tissue. Although Cd8 and Cd4 expression was upregulated in both O-HSV11 and O-HSV1211 treated group, that the IFN-γ production in O-HSV1211 treated-group was much higher than that in O-HSV1, O-HSV11 and O-HSV12 treated-group (Fig. 5C). These data implied that the O-HSV1211 can be used as the most potential therapeutic agent for tumor treatment. To better understand this observed tumor-control advantage, we evaluated the modulatory mechanism of O-HSV1211 in TME. Tumor-bearing mice were treated with O-HSV1 and O-HSV1211 on Days 7, 10, and 13 and euthanized on Day 21. O-HSV1211 effectively inhibited tumor growth and weights after intratumoral injection of viruses compared with that of PBS control or O-HSV1 (Fig. 5D and E). We further monitored the number of CD8+ and CD4+ T cells in tumors by flow cytometry and found that CD8+ and CD4+ T cell infiltration was increased after oncolytic virus treatment. Notably, the numbers of CD8 and CD4 T cells were further elevated in IL12 and CXCL11-armed oncolytic virus treated group (Fig. 5F).
Fig. 5.
IL12- and CXCL11-armed O-HSV1211 increased the efficiency of the oncolytic effect. A Schematic diagram of the oncolytic virus treatment. Sex and age matched C57BL/6J mice were implanted with MC38 cells (1×105 cells) on Day0, and oncolytic viruses or PBS were treated intratumorally on Days 7, 10 and 13. B The survival of tumor-bearing mice intratumorally treated with different oncolytic viruses (O-HSV1, O-HSV11, O-HSV12 and O-HSV1211) on Days 7, 10 and 13 was plotted using the Kaplan‒Meier method. Kaplan‒Meier survival curves were analyzed by the log-rank test. C Tumor-bearing mice were treated with different oncolytic viruses (O-HSV1, O-HSV11, O-HSV12 and O-HSV1211) on Day 7. Tumor tissue were harvested on day 10, total RNA isolated, and the expression of genes (Cd8, Cd4 and Ifng) were analyzed by quantitative RT‒PCR. D–M Sex and age matched C57BL/6J mice were implanted with MC38 cells (1×105cells) on Day0, and O-HSV1, O-HSV1211 or PBS were administered intratumorally on Days 7, 10 and 13. D Tumor volumes were measured every 3rd day. E Tumors were weighted on Day 21 after inoculation. F–K Tumor-infiltrating immune cells were stained with fluorescence-conjugated antibodies and analyzed by flow cytometry. F Numbers of CD8+ and CD4+ T cells. G–H The expression of IFN-γ among CD8+ and CD4+ T cells. I–K The expression of IL-4, IL-17 and Foxp3 among CD4+ T cells. L Kaplan‒Meier survival curve analysis of tumor-bearing mice treated with oncolytic viruses. Survival data were calculated by the log-rank test. M Tumor growth kinetics of the PBS, O-HSV1 and O-HSV1211 treated group were measured every 3rd day in indicated days (PBS, O-HSV1 and O-HSV1211, n=6). Tumor volumes are presented as the mean values ± standard deviation and were analyzed by Student's t test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Similarly, IFN-γ production was significantly elevated in tumor-infiltrated CD8+ and CD4+ T cells treated with O-HSV1211 (Fig. 5G and H). Although the frequencies of Th2 and Th17 cells were increased and the frequency of Treg cells was decreased in the O-HSV1211- and O-HSV1-treated groups compared with the control group, the proportions were indistinct between the O-HSV1- and O-HSV1211-treated groups (Fig. 5I–K). Furthermore, O-HSV1211 treatment prolonged the survival of mice, as the median survival times were 39 days for the O-HSV1211-treated group and 30 and 24 days for the O-HSV1 and control groups, respectively (Fig. 5L). The change in tumor volume was tracked in posttreated mice, and the IL12- and CXCL11-loaded oncolytic viruses delayed the time of tumor exacerbation (Fig. 5M).
Taken together, CXCL11 mediated the migration of CTL and Th1 cells into the TME, and IL12 promoted the production of IFN-γ in T cells. The combination of CXCL11 and IL12 dramatically augments IFN-γ production by CTLs and Th1 cells in the TME, which implicates the antitumor effect of O-HSV1211 associated with IFN-γ.
3.6. Blocking of IFN-γ abrogates the antitumor effect of O-HSV1211
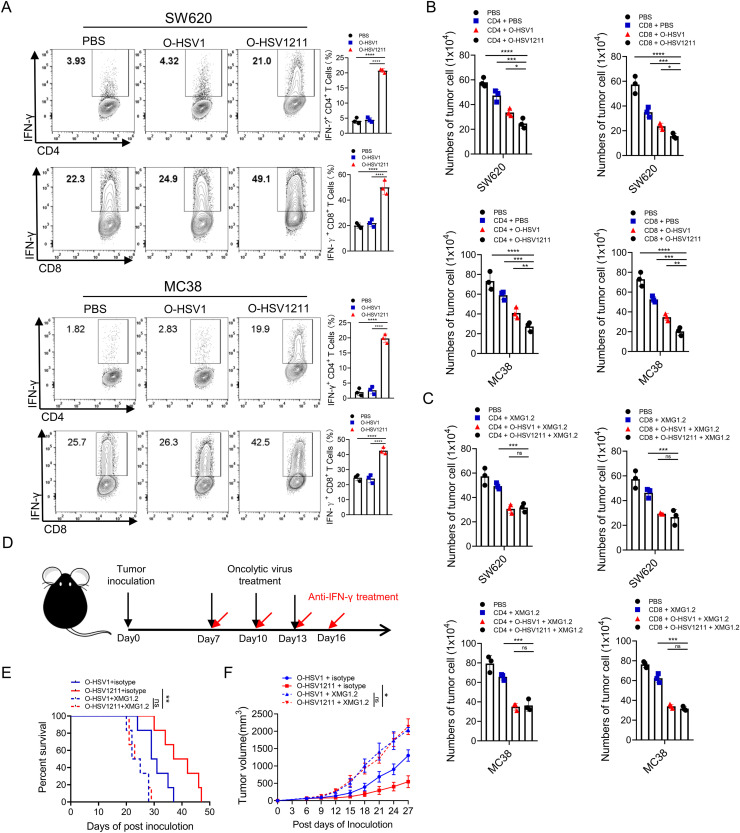
The duration of CXCL11 and IL12 release by O-HSV1211 in the TME results in increased IFN-γ, which derived from cytotoxic lymphocytes can directly enhance their cytotoxicity (Bhat et al., 2017; Curtsinger et al., 2012; Takeda et al., 2017). To answer the question of whether the enhanced antitumor phenotype is dependent on increased IFN-γ production in the case of O-HSV1211 treatment, we used an in vitro coculture model to evaluate the effect of O-HSV1211 on the cytotoxicity of CD8+ and CD4+ T cells. SW620 cells and MC38 cells were first cultured in plates precoated with anti-CD3 and anti-CD28 antibodies and then infected with different oncolytic viruses. Next, activated CD8+ and CD4+ T cells were cocultured with infected SW620 and MC38 cells, and the IFN-γ level was markedly increased in the O-HSV1211 infected group (Fig. 6A). The reduction in cell numbers further indicated the reinforced cytotoxicity of CD8+ and CD4+ T cells by O-HSV1211 compared with O-HSV1 (Fig. 6B). We further used a neutralizing anti-IFN-γ mAb (supplemented with 10 μg/ml XMG1.2 in medium) to block IFN-γ, and found that the tumor cell numbers were not significantly (ns) changed in the O-HSV1 and O-HSV1211 treated wells (Fig. 6C). Consistent with the in vitro results, neutralization of IFN-γ in the O-HSV1- and O-HSV1211-treated MC38 tumor models occurred on Days 7, 10, 13 and 16, and O-HSV1 and O-HSV1211 plus isotype were used as controls (Fig. 6D). Compared with that in the control group, the survival of mice was monitored after treatments, and the median survival was shortened from 31(O-HSV1 plus isotype) and 40 (O-HSV1211 plus isotype) days to 24 (O-HSV1 plus XMG1.2) and 25 (O-HSV1211 plus XMG1.2) days, respectively (Fig. 6E). We followed the tumor size in posttreated mice and observed tumor relapse earlier in both anti-IFN-γ-treated group (Fig. 6F). These data indicated that the antitumor effect of O-HSV1211 highly relied on IFN-γ production by CD8+ and CD4+ T cells.
Fig. 6.
Promoting the type 1 immune response of O-HSV1211 enhanced antitumor function. A–C Coculture of tumor cells with CD4+ or CD8+ T cells. Tumor cells were cultured in plates precoated with anti-CD3 and anti-CD28 antibodies for 48 h and infected with O-HSV1 and O-HSV1211 at an MOI of 1. CD4+ or CD8+ T cells were isolated and activated for 24 h and cocultured with infected SW620 or MC38 cells. A IFN-γ production in CD4+ and CD8+ T cells was analyzed by flow cytometry with a fluorescence-conjugated antibody. B Tumor cells were counted under a microscope (n=3). C Anti-IFN-γ (XMG1.2) antibody was used in the CD8+ and CD4+ T-cell coculture system to neutralize IFN-γ, and tumor cells were counted under a microscope (n=3). D Experimental schema. Sex and age matched C57BL/6J mice were implanted with MC38 tumor cells (1×105cells) on Day 0, and treated intratumorally with oncolytic viruses or PBS on Days 7, 10 and 13 (black arrows). Anti-IFN-γ antibody (5 mg/kg) or isotype control IgG (5 mg/kg rat IgG) was injected intraperitoneally on Days 7, 10, 13 and 16 post tumor implantations (red arrows). E The survival of tumor-bearing mice treated with O-HSV1 or O-HSV1211 plus isotype or anti-IFN-γ antibody was plotted using the Kaplan–Meier method. F Tumor growth in experiment E was measured every 3rd day (n=6). Tumor volumes are presented as the mean values ± standard deviation and analyzed by Student's t test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
Overall, these data demonstrated that the combination of IL12 and CXCL11 in an oncolytic virus achieved maximal survival of tumor-bearing mice and that the O-HSV1211 antitumor response is IFN-γ dependent.
4. Discussion
This work exploited the CRISPR/Cas9 system to edit the HSV-1 genome and demonstrated that recombinant HSV-1 can be obtained in less than 3 weeks. We engineered an oncolytic virus, O-HSV1211 armed IL12 and CXCL11, with the assistance of the CRISPR/Cas9 system, which has shown great promise in CRC treatment.
Typically, the HSV-1 genome must be incorporated into bacterial artificial chromosomes (BACs) for recombination in E. coli, and after the intended modification is screened, the viral genome is extracted and packed as a viral particle (Gierasch et al., 2006). In our report, CRISPR/Cas9-mediated gene editing provides a site-specific modification of the viral genome. Moreover, the adoption of CRISPR/Cas9 for the construction of recombinant virus significantly improved the efficiency in obtaining foreign armed viral clones. This approach will facilitate the study of oncolytic viruses for cancer treatment.
Oncolytic viruses have demonstrated evidence of efficacy (Ramelyte et al., 2021). The combination of oHSV with deletion of ICP34.5 and ICP47 in HSV-1 and immune checkpoint modulators (anti-CTLA4 and anti-OX40 antibodies) extended the lifespan of pancreatic ductal adenocarcinoma (PDAC)-bearing mice (Zhang et al., 2021). Oncolytic HSV expressing IL-12 with ICIs, including anti-CTLA4 and anti-PD1 antibodies, could extend the survival of glioma-bearing mice. Extensive studies have indicated that the tumor killing ability of oncolytic viruses alone is limited, while oncolytic viruses expressing immunomodulatory transgenes could further enhance the efficacy. For example, T-Vec delivers the GM-CSF gene, which makes a joint effort in the treatment of melanoma (Harrington et al., 2015). Oncolytic viruses expressing immunomodulatory genes, such as IL-12, can functionally induce antigen-specific CD8+ T-cell immune responses (Thomas et al., 2016). To specifically deliver the oncolytic virus to cancer, the binding site region of glycoproteins such gD and gH was replaced with a tumor-targeting single chain antibody scFv. Genetically modified HSVs have been successfully retargeted to many receptors, such as IL-13α2R, uPAR, and HER2, which improved the targeted killing of tumor cells (Gambini et al., 2012; Kamiyama et al., 2006; Zhang et al., 2017). An HSV-based oncolytic virus encoding a PD-1 antibody to block PD-1 has shown a significant antitumor response in glioblastoma (Passaro et al., 2019).
Since tumors have a complex immunosuppressive TME, a single therapeutic gene armed with an oncolytic virus may be unable to meet the requirement of antitumor activity. On this basis, oncolytic viruses delivering multiple immunoregulators will be a more potent approach to enhance immune cell-mediated antitumor immune responses. However, the traditional method of HSV viral genome editing is laborious and time-consuming. Although preliminary research has been reported for fluorescent gene insertion in the HSV genome with the CRISPR/Cas9 system, engineering HSV-1 requires more efficient approaches to modify the viral genome. We used the CRISPR/Cas9 system to delete the ICP34.5 and ICP47 genes and inserted a foreign gene into the ICP34.5 gene region in the HSV-1 genome. Our approach facilitates the development of multiple gene-armed oncolytic herpes simplex virus, and IL12/CXCL11-armed O-HSV1211 has been demonstrated to have enormous potential in CRC treatment. Moreover, O-HSV1 can be used as a basic oncolytic virus backbone for further insertion of different gene(s) in the ICP34.5 gene or other intergenic regions, such as UL26/UL27 and UL3/UL4.
5. Conclusions
In this report, the CRISPR/Cas9 system was successfully applied to HSV-1 viral genome manipulation for oncolytic viruses. We further identified that the dual gene (IL12 and CXCL11)-armed oncolytic virus can synergize to improve therapeutic outcomes in colon cancer. Application of the CRISPR/Cas9 system is simple and economical, allowing further investigation of diverse immunostimulatory cytokines and oncolytic viruses equipped with three or more genes as potential drugs for cancer treatments. The simplified and efficient method for HSV-1 genome editing for oncolytic purposes may also be applied to modify other dsDNA virus-based agents, such as adenoviruses.
Data availability
The authors declare that the data supporting the findings of this study are available within the article or upon request to the corresponding authors.
Ethics statement
For mice experiments, the animal care and experimental procedures are adhered to the protocol approved by the Institutional Animal Care and Use Committee of Nankai University.
CRediT authorship contribution statement
Nianchao Zhang: Data curation, Software, Investigation, Writing – review & editing, Writing – original draft. Jie Li: Data curation, Software, Writing – review & editing. Jingxuan Yu: Data curation, Software, Writing – review & editing. Yajuan Wan: Software. Cuizhu Zhang: Data curation, Software. Hongkai Zhang: Conceptualization, Methodology, Writing – review & editing. Youjia Cao: Conceptualization, Methodology, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81672010, and 81661148051 to YC). This work is also in the Programme of Introducing Talents of Discipline to Universities (B08011).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198979.
Contributor Information
Hongkai Zhang, Email: hkzhangnankai@163.com.
Youjia Cao, Email: yjcnankai@163.com.
Appendix. Supplementary materials
References
- Agarwalla P.K., Aghi M.K. Oncolytic herpes simplex virus engineering and preparation. Methods Mol. Biol. 2012;797:1–19. doi: 10.1007/978-1-61779-340-0_1. [DOI] [PubMed] [Google Scholar]
- Apetoh L., Quintana F.J., Pot C., Joller N., Xiao S., Kumar D., Burns E.J., Sherr D.H., Weiner H.L., Kuchroo V.K. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 2010;11(9):854–861. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat P., Leggatt G., Waterhouse N., Frazer I.H. Interferon-gamma derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell. Death Dis. 2017;8(6):e2836. doi: 10.1038/cddis.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommareddy P.K., Patel A., Hossain S., Kaufman H.L. Talimogene Laherparepvec (T-VEC) and other oncolytic viruses for the treatment of melanoma. Am. J. Clin. Dermatol. 2016;18(1):1–15. doi: 10.1007/s40257-016-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro F., Cardoso A.P., Goncalves R.M., Serre K., Oliveira M.J. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front. Immunol. 2018;9:847. doi: 10.3389/fimmu.2018.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G., Brett M.E., He B. Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the 134.5 protein of herpes simplex virus type 1. J. Virol. 2002;76(18):9434–9445. doi: 10.1128/JVI.76.18.9434-9445.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtsinger J.M., Agarwal P., Lins D.C., Mescher M.F. Autocrine IFN-gamma promotes naive CD8 T cell differentiation and synergizes with IFN-alpha to stimulate strong function. J. Immunol. 2012;189(2):659–668. doi: 10.4049/jimmunol.1102727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara H., Ino Y., Todo T. Oncolytic virus therapy: a new era of cancer treatment at dawn. Cancer Sci. 2016;107(10):1373–1379. doi: 10.1111/cas.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambini E., Reisoli E., Appolloni I., Gatta V., Campadelli-Fiume G., Menotti L., Malatesta P. Replication-competent herpes simplex virus retargeted to HER2 as therapy for high-grade glioma. Mol. Ther. 2012;20(5):994–1001. doi: 10.1038/mt.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh K., Stadler Z.K., Cercek A., Mendelsohn R.B., Shia J., Segal N.H., Diaz L.A., Jr. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019;16(6):361–375. doi: 10.1038/s41575-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S.J., Bi Y., Sun L., Gao D., Ding C., Li Z., Li Y., Cun W., Li Q. High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. PLoS Pathog. 2014;10(5) doi: 10.1371/journal.ppat.1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierasch W.W., Zimmerman D.L., Ward S.L., VanHeyningen T.K., Romine J.D., Leib D.A. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods. 2006;135(2):197–206. doi: 10.1016/j.jviromet.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Harrington K.J., Hingorani M., Tanay M.A., Hickey J., Bhide S.A., Clarke P.M., Renouf L.C., Thway K., Sibtain A., McNeish I.A., Newbold K.L., Goldsweig H., Coffin R., Nutting C.M. Phase I/II study of oncolytic HSVGM-CSF in combination with radiotherapy and cisplatin in untreated stage III/IV squamous cell cancer of the head and neck. Clin. Cancer Res. 2010;16(15):4005–4015. doi: 10.1158/1078-0432.CCR-10-0196. [DOI] [PubMed] [Google Scholar]
- Harrington K.J., Puzanov I., Hecht J.R., Hodi F.S., Szabo Z., Murugappan S., Kaufman H.L. Clinical development of talimogene laherparepvec (T-VEC): a modified herpes simplex virus type-1-derived oncolytic immunotherapy. Expert Rev. Anticancer Ther. 2015;15(12):1389–1403. doi: 10.1586/14737140.2015.1115725. [DOI] [PubMed] [Google Scholar]
- He B., Chou J., Brandimarti R., Mohr I., Gluzman Y., Roizman1 B. Suppression of the phenotype of gamma(1)34.5- herpes simplex virus 1: failure of activated RNA-dependent protein kinase to shut off protein synthesis is associated with a deletion in the domain of the alpha47 gene. J. Virol. 1997;71(8):6049–6054. doi: 10.1128/jvi.71.8.6049-6054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., Cradick T.J., Marraffini L.A., Bao G., Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Yan Z., Ma Y., Cao Y., He B. A Herpesvirus virulence factor inhibits dendritic cell maturation through protein phosphatase 1 and I B kinase. J. Virol. 2011;85(7):3397–3407. doi: 10.1128/JVI.02373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama H., Zhou G., Roizman B. Herpes simplex virus 1 recombinant virions exhibiting the amino terminal fragment of urokinase-type plasminogen activator can enter cells via the cognate receptor. Gene Ther. 2006;13(7):621–629. doi: 10.1038/sj.gt.3302685. [DOI] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M., Kuchroo V.K. IL-17 and Th17 Cells. Ann. Rev. Immunol. 2009;27(1):485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Lee Y.L., Ye Y.L., Yu C.I., Wu Y.L., Lai Y.L., Ku P.H., Hong R.L., Chiang B.L. Construction of single-chain interleukin-12 DNA plasmid to treat airway hyperresponsiveness in an animal model of asthma. Human Gene Ther. 2001;12(17):2065–2079. doi: 10.1089/10430340152677412. [DOI] [PubMed] [Google Scholar]
- Li Y., Zhang C., Chen X., Yu J., Wang Y., Yang Y., Du M., Jin H., Ma Y., He B., Cao Y. ICP34.5 protein of herpes simplex virus facilitates the initiation of protein translation by bridging Eukaryotic Initiation Factor 2α (eIF2α) and protein phosphatase 1. J. Biol. Chem. 2011;286(28):24785–24792. doi: 10.1074/jbc.M111.232439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Li H., Hao M., Xiong D., Luo Y., Huang C., Yuan Q., Zhang J., Xia N. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing of HSV-1 virus in human cells. Sci. Rep. 2016;6(1) doi: 10.1038/srep34531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.L., Robinson M., Han Z.Q., Branston R.H., English C., Reay P., McGrath Y., Thomas S.K., Thornton M., Bullock P., Love C.A., Coffin R.S. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 2003;10(4):292–303. doi: 10.1038/sj.gt.3301885. [DOI] [PubMed] [Google Scholar]
- Liu Z., Ravindranathan R., Li J., Kalinski P., Guo Z.S., Bartlett D.L. CXCL11-Armed oncolytic poxvirus elicits potent antitumor immunity and shows enhanced therapeutic efficacy. Oncoimmunology. 2016;5(3) doi: 10.1080/2162402X.2015.1091554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R., Sarisky R.T., Weber P.C., Weller S.K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschulla T., Berry R., Gerke C., Doring M., Busch J., Paijo J., Kalinke U., Momburg F., Hengel H., Halenius A. A highly conserved sequence of the viral TAP inhibitor ICP47 is required for freezing of the peptide transport cycle. Sci. Rep. 2017;7(1):2933. doi: 10.1038/s41598-017-02994-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineta T., Rabkin S.D., Yazaki T., Martuza W.D.H.R.L. Attenuated multi–mutated herpes simplex virus–1 for the treatment of malignant gliomas. Nat. Med. 1995;1:938–943. doi: 10.1038/nm0995-938. [DOI] [PubMed] [Google Scholar]
- Pan S., Liua X., Ma Y., Cao Y., He B. Herpes simplex virus 1 γ134.5 protein inhibits STING activation that restricts viral replication. J. Virol. 2019;92(20):e01015–18. doi: 10.1128/JVI.01015-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passaro C., Alayo Q., De Laura I., McNulty J., Grauwet K., Ito H., Bhaskaran V., Mineo M., Lawler S.E., Shah K., Speranza M.C., Goins W., McLaughlin E., Fernandez S., Reardon D.A., Freeman G.J., Chiocca E.A., Nakashima H. Arming an oncolytic herpes simplex virus type 1 with a single-chain fragment variable antibody against PD-1 for experimental glioblastoma therapy. Clin. Cancer Res. 2019;25(1):290–299. doi: 10.1158/1078-0432.CCR-18-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppers J., Mulvey M., Khoo D., Mohr I. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J. Virol. 2000;74(23):11215–11221. doi: 10.1128/jvi.74.23.11215-11221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post1 D.E., Fulci2 G., Chiocca E.A., Meir E.G.V. Replicative oncolytic herpes simplex viruses in combination cancer therapies. Curr. Gene Ther. 2004;4(1):41–51. doi: 10.2174/1566523044577988. [DOI] [PubMed] [Google Scholar]
- Raafat N., Sadowski-Cron C., Mengus C., Heberer M., Spagnoli G.C., Zajac P. Preventing vaccinia virus class-I epitopes presentation by HSV-ICP47 enhances the immunogenicity of a TAP-independent cancer vaccine epitope. Int. J. Cancer. 2012;131(5):E659–E669. doi: 10.1002/ijc.27362. [DOI] [PubMed] [Google Scholar]
- Ramelyte E., Tastanova A., Balazs Z., Ignatova D., Turko P., Menzel U., Guenova E., Beisel C., Krauthammer M., Levesque M.P., Dummer R. Oncolytic virotherapy-mediated anti-tumor response: a single-cell perspective. Cancer Cell. 2021;39(3):394–406. doi: 10.1016/j.ccell.2020.12.022. e394. [DOI] [PubMed] [Google Scholar]
- Russell T.A., Stefanovic T., Tscharke D.C. Engineering herpes simplex viruses by infection–transfection methods including recombination site targeting by CRISPR/Cas9 nucleases. J. Virol. Methods. 2015;213:18–25. doi: 10.1016/j.jviromet.2014.11.009. [DOI] [PubMed] [Google Scholar]
- Takeda K., Nakayama M., Hayakawa Y., Kojima Y., Ikeda H., Imai N., Ogasawara K., Okumura K., Thomas D.M., Smyth M.J. IFN-gamma is required for cytotoxic T cell-dependent cancer genome immunoediting. Nat. Commun. 2017;8:14607. doi: 10.1038/ncomms14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallóczy Z., Virgin I.V.H., Levine B. PKR-dependent xenophagic degradation of herpes simplex virus type 1. Autophagy. 2014;2(1):24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- Thomas E.D., Meza-Perez S., Bevis K.S., Randall T.D., Gillespie G.Y., Langford C., Alvarez R.D. IL-12 Expressing oncolytic herpes simplex virus promotes anti-tumor activity and immunologic control of metastatic ovarian cancer in mice. J. Ovarian Res. 2016;9(1):70. doi: 10.1186/s13048-016-0282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todo T., Martuza R.L., Rabkin S.D., Johnson P.A. Oncolytic herpes simplex virus vector with enhancedMHC class I presentation and tumor cell killing. PANS. 2001;98:6396–6401. doi: 10.1073/pnas.101136398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley R.J., Roizman B. Herpes simplex virus infections. Lancet. 2001;357(9267):1513–1518. doi: 10.1016/S0140-6736(00)04638-9. [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang W., Wang R., Zhang N., Shang H., Bi Y., Chen D., Zhang C., Li L., Yin J., Zhang H., Cao Y. Reshaping the immune microenvironment by oncolytic herpes simplex virus in murine pancreatic ductal adenocarcinoma. Mol. Ther. 2021;29(2):744–761. doi: 10.1016/j.ymthe.2020.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Takaku M., Zou L., Gu A.D., Chou W.C., Zhang G., Wu B., Kong Q., Thomas S.Y., Serody J.S., Chen X., Xu X., Wade P.A., Cook D.N., Ting J.P.Y., Wan Y.Y. Reversing SKI–SMAD4-mediated suppression is essential for TH17 cell differentiation. Nature. 2017;551(7678):105–109. doi: 10.1038/nature24283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Rajput A., Jin N., Wang J. Mechanisms of immunosuppression in colorectal cancer. Cancers (Basel) 2020;12(12) doi: 10.3390/cancers12123850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article or upon request to the corresponding authors.