Abstract
The potent liver carcinogen aflatoxin B1 (AFB1) is metabolized by cytochrome P450 to the mutagenic epoxide. We have observed that activated AFB1 also strongly induced mitotic recombination in the yeast Saccharomyces cerevisiae. To compare the recombinogenicity of AFB1 to its mutagenicity, three metabolically competent S. cerevisiae strains have been constructed. The frequencies of induced recombinants resulting from gene conversion or chromosomal translocations were determined by different prototrophic selections using two strains, whereas the inducibility of forward mutations was determined by the frequency of drug resistance in the third strain. Human cytochrome P4501A1- (CYP1A) and NADPH-cytochrome P450-oxidoreductase cDNAs were expressed in the strains to ensure intracellular metabolism to the epoxide. Exposure of the strains to AFB1 resulted in a 139- and 24-fold increase in the translocation and gene conversion frequencies, respectively, whereas the mutation frequency was increased only 3-fold. In contrast, benzo[a]pyrene-7,8-dihydrodiol and ethyl methanesulfonate induced mutation and mitotic recombination to similar degrees. We conclude that AFB1 exerted a strong recombinogenic, but only a weak mutagenic, effect. The recombinogenicity of AFB1 in yeast may indicate a mechanism for the high proportion of loss of heterozygosity that has been detected in AFB1-related human liver cancers.
INTRODUCTION
The mycotoxin AFB13 is a potent rodent carcinogen. It efficiently induces liver carcinomas in rats and rainbow trouts, but less so in mice (reviewed in Ref. 1). The mycotoxin is of special concern in certain geographic areas in East Asia and Southern Africa, where AFB1 contamination of stocked food is a major problem due to particular climatic conditions. Epidemiological data have provided strong evidence for an association of AFB1 exposure and human liver cancer (for review, see Ref. 2). Partly for these reasons, AFB1 has been classified by the IARC as a class 1 human carcinogen (3).
AFB1 is metabolized by cytochrome P450-dependent mono-oxygenases, mostly by CYP1A2 and CYP3A4, to the mutagenic but highly unstable AFB1-8,9-exo-epoxide (activated AFB1; reviewed in Ref. 1). AFB1 can also be efficiently activated by CYP1A1 to a genotoxic (4-6) or cytotoxic metabolite (7-9). The epoxide form of AFB1 may be detoxified when inactivated by conjugation with glutathione, a pathway explaining the low susceptibility of mice toward AFB1 (10), or by reaction with microsomal epoxide hydrolase. Genetic variability of these two detoxification systems has been recognized to influence the susceptibility of humans to HCCs, and in individuals exposed to AFB1, higher levels of AFB1-serum albumin are correlated with the presence of an epoxide hydrolase allele (EPHX2), as well as with a glutathione-transferase null allele (GSTM1; Ref. 11).
Binding of activated AFB1 to the N7 position of guanine results in the formation of the unstable trans-8,9-dihydro-(N7-guanyl)-9-hydroxy-aflatoxin B1 adduct, which may undergo a hydrolytic ring opening reaction to result in the more stable formamidopyrimidine form (12). A direct consequence of DNA binding in both prokaryotic and eukaryotic organisms is the introduction of point mutations. In bacteria, activated AFB1 generates frameshift (13) and missense mutations (14), mostly G to T transversions (15-17). G to T transversions are also the major class of mutations found in both the supF target gene, when a shuttle plasmid was incubated in vitro with metabolically activated AFB1 and transfected into human Ad293 cells (18, 19), and in the HPRT gene, when a human lymphoblastoid cell line expressing CYP1A1 was exposed to AFB1 (20). Hot spots for AFB1 mutations include six contiguous G residues in the HPRT gene (20) and the third position in codon 249 of the human p53 tumor suppressor gene; the latter case was observed in liver cancers isolated from patients living in AFB1-contaminated areas (21-27), but not in AFB1-unrelated liver cancers (for review, see Ref. 28). Moreover, the same codon served as a hot spot when human hepatocytes were exposed to activated AFB1 in vitro (29).
Although the carcinogenicity and mutagenicity of AFB1 are thus correlated, other genotoxic mechanisms, such as recombination, may also be important in inducing liver cancer. For example, LOH is postulated to result in homozygosis of recessive mutations in tumor-suppressing genes, such as p53, and thus confer a greater proliferative ability. LOH may result from gene conversion, mitotic crossing-over (reciprocal exchange), chromosome loss, and/or duplication. Mutations in p53 also confer greater genetic instability, as illustrated in the enhanced frequencies of translocations in p53 knockout mice (30) and the enhanced frequencies of gene amplification in p53-deficient cell lines (31, 32). The ability of activated AFB1 to promote sister chromatid exchanges in V79 Chinese hamster cells (33) and intrachromosomal recombination between a 2-kb duplication of the HPRT gene in SP5/V79 cells (34) underscores the importance of determining the types of recombination events that can be stimulated by AFB1.
Mitotic, homologous recombination events include gene conversion or nonreciprocal exchange and reciprocal exchange. Because the former may not be associated with exchange of flanking markers and not generate chromosome rearrangements (35), whereas the latter may result in inversions, deletions, and translocations, assay systems that monitor reciprocal exchange and gene conversion separately are useful. Strains of Saccharomyces cerevisiae that independently monitor gene conversion events, translocations, and deletions (36-38) have been constructed to quantify the recombinogenicity of DNA damaging agents (39, 40). Using a strain construction to monitor allelic recombination, we have demonstrated previously that exposure to AFB1 can stimulate gene conversion in yeast (41). In this study, we demonstrate that AFB1 can also stimulate the formation of directed reciprocal translocations (reciprocal exchange) in yeast, thus underscoring the recombinogenicity of AFB1. In comparing the recombinogenicity and mutagenicity of AFB1 with other known carcinogens, we suggest that the recombinogenicity of AFB1 may contribute to its strong carcinogenicity.
MATERIALS AND METHODS
Chemicals.
BaP-7,8-DHD was purchased from Midwest Research Institute (Kansas City, MI) and AFB1 from Fluka (Buchs, Switzerland). Both chemicals were dissolved in DMSO. EMS was obtained from Eastman Kodak (Rochester, NY). Amino acids, Ade, and Ura were purchased from Merck (Dietikon, Switzerland), yeast nitrogen base and bacto agar from Difco (Chemie Brunschwig, Basel, Switzerland). DNA modifying enzymes were from Boehringer (Rotkreuz, Switzerland).
Strains and Media.
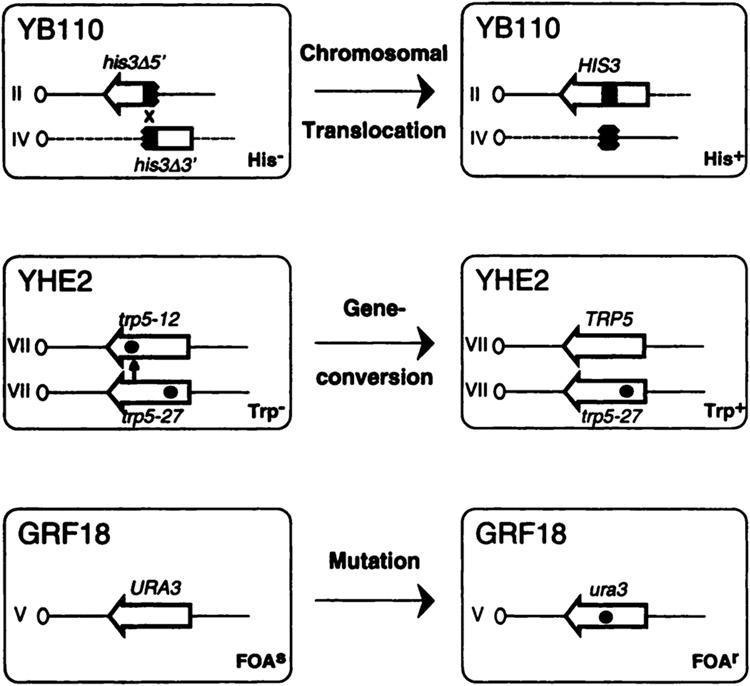
Yeast transformants were grown on YM medium (0.67% yeast nitrogen base without amino acids, 2% glucose) containing, if required, the following supplements: Ade, Ura, Trp, and His at 20 μg/ml, Ile and Leu at 30 μg/ml, and Val at 150 μg/ml. Strains YHE2 (MATa/MATα, ade2-40/ade2-119, trp5-12/trp5-27, ilvl-92/ilvl-92, ura3Δ5/ura3Δ5 (42) and YB110 (MATa/MATα, ade2-101/ade2-101, ura3-52/ura3-52, his3-Δ200/his3-Δ200, trpl-Δ1/trpl-Δ1, leu2/LEU2, GAL1::his3-Δ5′GAL1, trpl::his3-Δ3′/trpl-Δ1, LYS2/lys2-801; Ref. 43) were used to quantitate allelic gene conversion and chromosomal translocation, respectively. The haploid strain GRF18 (MATα, his3-11, 15, leu2-3, 112, canR)4 was used to monitor mutations in URA3. YPD medium contained 1% yeast extract, 2% bacto peptone, and 2% glucose.
Plasmids.
All DNA manipulations were performed according to standard protocols (44). Plasmids pSB229 (45) and pCS316 (4) contain the 2-μm origin of replication, URA3, hOR, and either CYP1A1 or CYP1A2 cDNAs, respectively. The cDNAs are expressed under control of the constitutive glyceraldehyde phosphate dehydrogenase promoter. The parental vectors pDP34 (46) and pNW144 (47) have been described previously. Plasmids pCS508 and pCS512 are derived from pNW144 and pSB229, respectively, in which the LEU2 marker substitutes for URA3. For these manipulations, the 2.2-kb XhoI/SalI fragment from plasmid YEp13 (48) containing LEU2 was isolated from an agarose gel using Geneclean (BIO 101 Inc., Vista, CA), and the ends were converted to blunt ends. The fragment was inserted into the blunt-ended HindIII site of pUC19 (49), and the SacI/PstI fragment containing the vector and LEU2 sequences was isolated and used to replace the SacI/PstI fragment in pDP34, thus resulting in plasmid pCS508. A 4-kb NruI/ScaI fragment from pCS508 containing LEU2 and part of the ampicillin resistance gene was isolated and ligated with the large NruI/ScaI fragment of pSB229 to give pCS512. These manipulations resulted in the replacement of URA3 by LEU2 but left the two expression cassettes in pSB229 unaffected. All plasmids were transformed into yeast according to standard protocols (50).
Determination of Enzyme Activity.
EROD activities were determined from exponentially growing cells (A600 = 0.8–1.5). Cells were resuspended at a density of 7 A600 units/2 ml in 0.1 m Tris buffer (pH 7.8), and 7-ethoxyresorufin was added to 1 μM. The reaction was started by adding NADPH to 0.25 μM, and resorufin formation was recorded using a Perkin-Elmer MPF-3L fluorescence spectrophotometer according to Rutten et al. (51). Resorufin (50 pmol) was added as an internal standard.
Genotoxicity Tests.
Exponentially growing yeast cells at a cell density of 108 cells/ml were exposed to chemicals in 1 ml of 0.1 m sodium phosphate buffer (pH 7.5) for 4 h at 30°C, unless otherwise indicated. The cells were then pelleted in a clinical centrifuge, washed, and diluted in supplemented minimal medium. For gene conversion assays, YHE2 transformants were grown in YM Ade-Ile-Trp-Leu-Val. After exposure to mutagen, 50 μl were directly plated on YM Ade-Ile-Leu-Val-Ura plates containing 2% bacto agar to select for Trp+ clones and 100 μl of a 10−4 dilution on full medium YPD plates (52) to plate for survivors. Plates were incubated for 3 and 2 days, respectively, at 30°C. For translocation assays, YB110 transformants were grown in YM His-Ade-Trp. After mutagen treatment, cells were resuspended at a density of 4 × 108/ml, and 250 μl were plated directly on YM Ade-Ura-Trp to select for His+ recombinants and 100 μl of a 10−5 dilution were plated on YPD for survivors. Selection plates were incubated at 30°C and scored after 7 days. For mutation assays, GRF18 transformants were grown in YM His and exposed to mutagen in buffer at a density of 107 cells/ml. The cells were then washed with YPD, survivors were titrated on YPD plates, and 0.2 ml was used to inoculate 10 ml of YPD. The cultures were incubated on a 30°C shaker for 6–7 generations to allow expression of the Ura− phenotype. To select for ura3 mutants, 200-μl aliquots were plated on YM His-Leu-Ura plates supplemented with 1 mg/ml FOA (PCR Inc., Gainsville, FL), and to quantitate total survivors, 100-μl aliquots of a 10−4 dilution were plated on YPD. Plates were incubated at 30°C, and FOA and YPD plates were scored after 3 and 2 days, respectively. To select for α-aminoadipate-resistant lys2 mutants, 200-μl aliquots were plated on YM His-Leu plates supplemented with 30 μg/ml L-lysine and 2 mg/ml α-aminoadipic acid (Sigma, Buchs, Switzerland), which was solved in H2O by adjusting the pH to 6 with KOH. Plates were incubated at 30°C and scored after 3 days.
Characterization of Chromosome Rearrangements by CHEF.
Chromosomal DNA was prepared from AFB1-induced recombinants as described previously (37). CHEF gels (35) were run at 6 V/cm for 26 h. Gels were subsequently stained with ethidium bromide, and translocations were visually identified.
RESULTS
AFB1 Induces Directed Chromosomal Translocations in S. Cerevisiae.
It was observed previously that the mycotoxin AFB1, when intracellularly activated by human cytochrome P450 enzymes from family 1, strongly induced mitotic gene conversion events between two trp5 alleles in S. cerevisiae (41). In this study, we asked whether metabolically activated AFB1 would also increase the recombination frequency of short homologous sequences located on nonhomologous chromosomes. To address this question, the plasmids pSB229 (45), pCS316 (4), or the vector pNW144 (47) were introduced into the diploid S. cerevisiae strain YB110 (43) by selecting for Ura+ transformants (Table 1). This His− strain contains two his3 fragments, truncated at either the 5′ or 3′ end, that are stably integrated in chromosomes II and IV, respectively (Fig. 1). His+ prototrophs containing reciprocal translocations of the long arms of chromosomes II and IV are generated by mitotic recombination between 300 bp of shared homology (37). The presence of the individual plasmids in YB110 confers metabolic competence by directing the coexpression of human CYP1A1 (pSB229) or CYP1A2 (pCS316) cDNAs with a cDNA coding for hOR. In contrast, transformants of YB110 containing vector pNW144 do not express these human cDNAs.
Table 1.
Plasmids used in the study
| Plasmid | Expressed cDNAs | Selection marker |
|---|---|---|
| pSB229 | CYP1A1 + hOR | URA3 |
| pCS316 | CYP1A2 + hOR | URA3 |
| pCS512 | CYP1A1 + hOR | LEU2 |
| pDP34 | URA3 | |
| pNW144 | URA3 | |
| pCS508 | LEU2 |
Fig. 1.
Yeast strains designed to monitor recombination and mutagenesis. The diploid strain YB110 carries two truncated fragments of the his3 gene (broken arrow) on chromosomes II and IV. The 3′ end of the amino acid coding sequence for each gene is designated by an arrowhead. A recombination event involving ~300 bp of shared sequence homology (shaded area) results in the reconstitution of HIS3 and thus a selectable His+ phenotype. Dashed lines and straight lines represent chromosome IV and II sequences, respectively. Diploid strain YHE2 carries two nonfunctional trp5 alleles on chromosome VII homologues. Both alleles contain nonrevertible point mutations (black dots) at different locations. Mitotic gene conversion results when sequence information from the lower allele is nonreciprocally transferred (small vertical arrow) to the upper allele, thereby generating TRP5 and a selectable Trp+ phenotype. Haploid strain GRF18 carries a functional URA3 gene on chromosome V. The introduction of a mutation or deletion (black dot) inactivates the gene and confers resistance to FOA. The changes in phenotypes observed for the different strains are indicated.
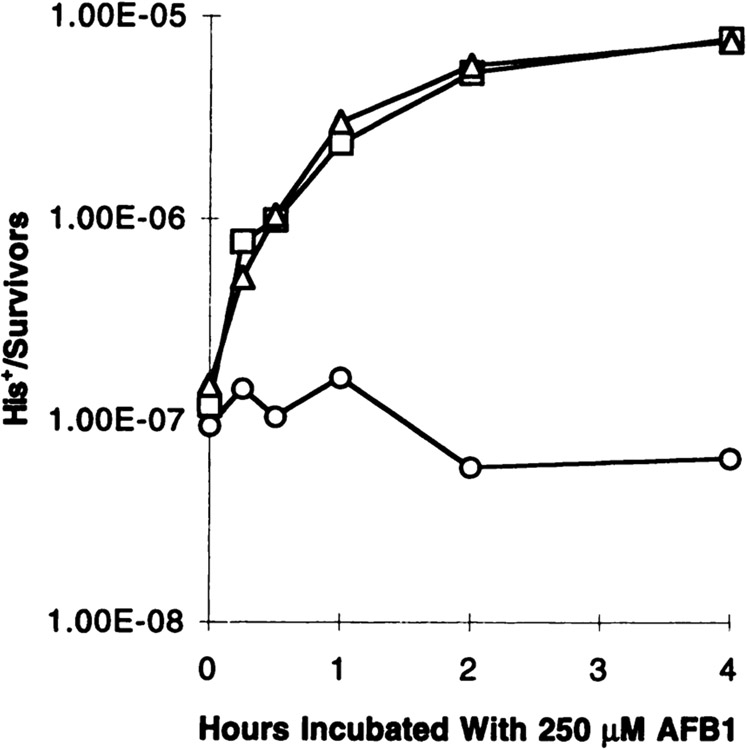
Activation of AFB1 by human enzymes present in YB110 transformants was verified by the stimulation of His+ recombinants after exposure to 250 μM of AFB1. Before AFB1 treatment, His+ recombinants occurred at very low frequencies (Fig. 2): (1.19 ± 0.43) × 10−7 for YB110pSB229, (1.49 ± 1.40) × 10−7 for YB110pCS316, and (0.94 ± 0.55) × 10−7 for YB110pNW144. With increasing time of exposure to AFB1, the recombination frequency steadily increased in strains YB110pSB229 and YB110pCS316 to 7.6 × 10−6, reaching a 51–64-fold increase over the background frequency. In contrast, the vector-transformed strain YB110pNW144 did not respond to AFB1 due to the lack of human CYP1A enzymes. At 4 h incubation with AFB1, the response curve reached a plateau in the two strains containing human enzymes, suggesting that no additional AFB1 was activated or recombination was saturated. The recombination frequencies were consistent with previous observations that both human cytochrome P450 enzymes from family 1 are able to activate AFB1 to a genotoxic product (4). Therefore, an incubation time of 4 h was chosen for further experiments.
Fig. 2.
AFB1 significantly induces reciprocal translocations in metabolically competent YB110 transformants. YB110 cells expressing CYP1A1 (□) or CYP1A2 (Δ), in combination with hOR or expressing no human cDNA (○), were exposed to 250 μM AFB1 at indicated times. His+ recombinants and survivors were quantitated on appropriate media (see “Materials and Methods”). Translocation frequencies are given as the ratio of His+ clones per survivors.
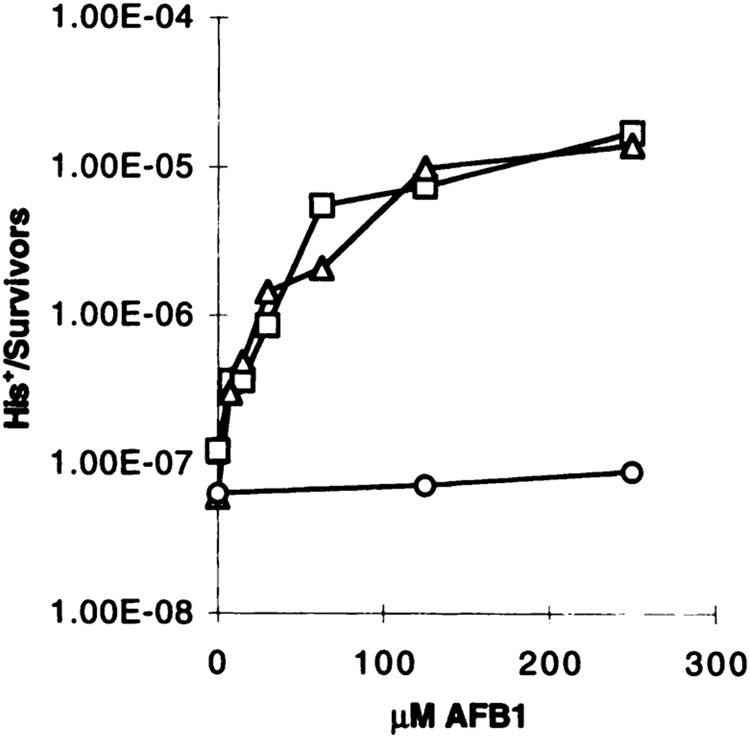
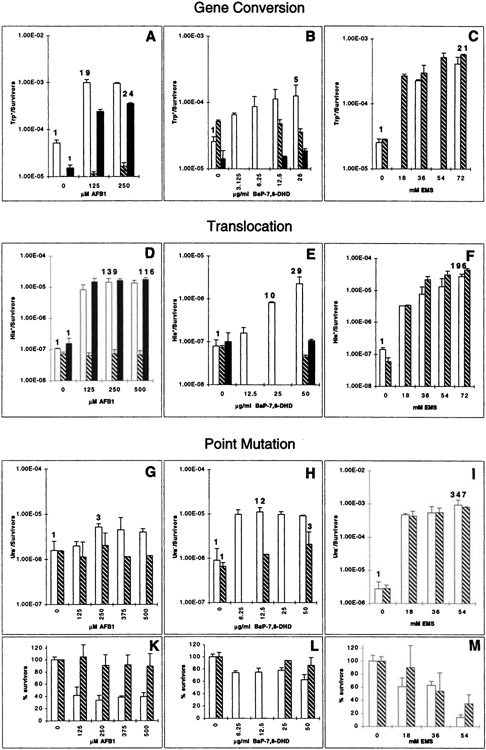
The increase in recombination frequencies in strains YB110pSB229 and YB110pCS316 after AFB1 treatment was dose dependent. After exposure for 4 h to different doses of AFB1, both metabolically competent strains exhibited a similar dose-response curve with a steep increase at low AFB1 doses and a saturation in the range of 250 μM (Fig. 3). Higher doses of AFB1 resulted in a lower recombination frequency, a behavior consistent with previously observed AFB1-induced gene conversion (data not shown). The exposure of the strains to different doses of AFB1 was repeated in at least four independent experiments, and the His+ recombination frequencies were calculated (Fig. 4D). Consistent with our previous findings, a significant increase in the His+ recombination frequency was observed in both metabolically competent strains, whereas the control strain expressing no human cDNA was not responsive. At 250 μM AFB1, the recombination frequency was increased by a factor of 139 in the CYP1A1-expressing strain, and in the CYP1A2-expressing strain by a factor of 116 at 500 μM AFB1. Thus, the level of induction and dose response are reproducible. The induction of recombination apparently occurred in the absence of cytotoxicity. Even at the highest dose applied, more than 83% of the cells survived the treatment (data not shown). From this, it was concluded that the AFB1-stimulated recombination did not result from the secondary unrelated effects of dying cells.
Fig. 3.
Dose-dependent increase in His+ recombinants when YB110 transformants expressing P450 enzymes are exposed to AFB1. Cells were exposed for 4 h to different doses of AFB1, and frequencies of His+ recombinants were calculated. The symbols are the same as in Fig. 2.
Fig. 4.
Panels show the summary of gene conversion, translocation, and point mutation data. The different genotoxic end points that are detected with the individual strains are indicated and shown for AFB1 (A, D, and G), BaP-7,8-DHD (B, E, and H), and EMS (C, F, and I). Strain YHE2 (A–C-), YB110 (D–F), or GRF18 (G–M) expressing CYP1A1 and hOR (white columns), CYP1A2 and hOR (black columns), or expressing no human cDNA (striped columns) were exposed to chemicals as described in “Materials and Methods.” The numbers above selected columns give the fold induction over the background frequency. Data in A were taken from Sengstag and Würgler (41). The means and SDs were determined from four to five experiments performed in parallel (B and D), from three experiments performed in parallel (E, G, and I), or from experiments performed in duplicate (A, C, F, and H). Cytotoxicity data are shown only for the mutation tester strain GRF18 (K–M). With both of the other strains, YB110 and YHE2, survival rates were all above 65% when gene conversion or translocation was induced by AFB1 or BaP-7,8-DHD, and above 54% when EMS was applied (not shown). Bars, SD.
The electrophoretic karyotypes of 10 spontaneous His+ recombinants from a YB110pNW144 transformant and 10 AFB1-induced His+ recombinants from a YB110pSB229 transformant were determined to verify that the induced recombinants were translocations and to ask whether unusual chromosomal rearrangements appeared among the AFB1-induced recombinants that did not appear among the spontaneous recombinants. In recombinants containing reciprocal translocations, HIS3 is located on CEN2::IV, whereas a his3 gene fragment lacking both 3′ and 5′ ends is located on the translocation designated as CEN4::II, as shown previously (37) and depicted in Fig. 1. Recombinants containing nonreciprocal translocations contain CEN2::IV, but not CEN4::II, and are viable in diploids that contain both wild-type copies of chromosomes II and IV. Ethidium bromide stains of CHEF gels revealed that, among the spontaneous recombinants, six recombinants were reciprocal translocations, one was a nonreciprocal translocation, and three recombinants were classified as “other” but had the wild-type configuration of chromosomes (data not shown). Among the induced recombinants, eight were reciprocal translocations, one was a nonreciprocal translocation, and one recombinant was classified as “other,” but had the wild-type configuration of chromosomes. Although the recombinants classified as “other” were not further investigated, this class has been studied previously and may include rare conversion events of the his3-Δ200 allele that does not result in translocation. Thus, reciprocal translocations predominate among both spontaneous and AFB1-induced recombinants, and no unusual types of recombination events were evident among the AFB1-induced recombinants that did not appear among the spontaneous recombinants.
The same doses of AFB1 also induced gene conversion between two trp5 alleles in strain YHE2. When transformants of strain YHE2, containing the plasmids pSB229 or pCS316, were exposed to the mycotoxin (Fig. 4A), a 19-fold increase was observed in the frequency of Trp+ recombinants in the strain expressing CYP1A1 and hOR, whereas a 24-fold increase over the spontaneous frequency (1–2 × 10−5 Trp+/survivors) was observed with the strain expressing CYP1A2 and hOR. At both AFB1 doses applied, survival of the CYP1A1-expressing strain was reduced to 65%, whereas no reduction in survival was observed with the other two strains (data not shown). From these results, it was concluded that the translocation tester strain YB110 responded with a higher sensitivity than the gene conversion tester strain YHE2 to AFB1. The control strain YHE2pDP34 did not respond to AFB1.
Induction of Chromosomal Translocations by Other Chemicals.
To test whether another carcinogen that depended on P450-mediated activation was able to induce chromosomal translocations in strain YB110, the transformed strain was exposed to the polycyclic aromatic hydrocarbon BaP-7,8-DHD. This chemical is metabolized by CYP1A1 to the ultimate carcinogen BaP-7,8-diol-9,10-epoxide (4, 53, 54). A dose-dependent increase in the translocation frequency was observed in strain YB110pSB229 expressing CYP1A1 and hOR (Fig. 4E), whereas the strain expressing CYP1A2 or no human enzyme was not responsive. At 50 μg/ml, a dose resulting in 83% survival of the cells (data not shown), the translocation frequency was elevated by a factor of 29 above background. The same compound also induced gene conversion in strain YHE2 (Fig. 4B), in which a maximal 5-fold induction over background was observed at a dose of 25 μg/ml. This dose resulted in 84% survival, whereas no reduction in viability was observed at lower doses or at all doses in experiments with the other two strains (data not shown). Although BaP-7,8-DHD induced both recombinogenic events, chromosomal translocation and gene conversion, the recombinogenicity of the mycotoxin AFB1 was greater than BaP-7,8-DHD in both strains. To determine whether the greater recombinogenicity of AFB1 also correlated with greater mutagenicity, the mutagenic activities of AFB1 and BaP-7,8-DHD were then tested in a haploid yeast strain.
Low Mutagenic Activity of Activated AFB1 in S. Cerevisiae.
To monitor the mutagenic activity of metabolically activated AFB1 in S. cerevisiae, frequencies of forward mutations in URA3 were determined by selecting for drug resistance using the antimetabolite FOA. This drug is cytotoxic in wild-type cells but not in ura3 mutants. Mutations in URA3 were studied in a haploid strain to ensure that URA3 was not inactivated by a recombination-mediated process between homologues. Metabolic competence was conferred to strain GRF18 by introducing plasmid pCS512, which is identical to plasmid pSB229, except that LEU2 substitutes for URA3 as a selective marker (Table 1). GRF18 transformants were therefore propagated on Leu-deficient medium. The control plasmid pCS508 was identical to the control plasmids pNW144 and pDP34, except that LEU2 substituted for URA3. Strains GRF18pCS512 and GRF18pCS508 were grown to exponential phase and exposed to AFB1 at different doses for 4 h, identical to the protocol using strains YHE2 or YB110. After washing the cells with buffer, cytotoxic effects were determined by plating an aliquot of the cells on complete medium (YPD). The washed cells were diluted (see “Materials and Methods”) and incubated for 6–7 generations in YPD to ensure expression of the Ura− phenotype before plating on FOA plates. The titer of Ura− FOAr clones and surviving clones was then determined, and the mutation frequency (FOAr mutants/survivors) was calculated.
The same dose of AFB1 that induced a 129-fold increase in chromosomal translocation was able to increase the mutation frequency in strain GRF18pCS512 by 3-fold (Fig. 4G). This was paralleled by a reduction in cell viability to 35% (Fig. 4K), suggesting that AFB1 was able to enter the cell and that its metabolite exerted a cytotoxic effect. Thus, AFB1 exerted only a minor mutagenic effect in S. cerevisiae, whereas the same compound strongly stimulated mitotic recombination. As was expected, no genotoxic or cytotoxic effects were observed in the control strain GRF18pCS508 that does not express the human cDNAs (Fig. 4, D and G).
Because strain GRF18pCS512 contained a similar but nonidentical expression plasmid as strain YB110pSB229, the low degree of AFB1 mutagenicity in the former strain might be due to a lower expression level of CYP1A1 and/or hOR compared with the latter strain. Therefore, CYP1A1 -specific enzyme activity was determined in the three strains expressing CYP1A1 and hOR. Identically grown cultures of strains YHE2pSB229, YB110pSB229, and GRF18pCS512 were harvested during the exponential phase, and EROD activities were determined. The three strains did not significantly differ in their specific enzyme activities (Table 2), indicating that comparable levels of the human enzymes were present.
Table 2.
EROD activity of transformed yeast strains
| Strain | Expressed cDNAs | EROD activity (pmol resorufin formed/min × A600 units of cells)a |
|---|---|---|
| YHE2pSB229 | CYP1A1 + hOR | 36.3 ± 4.2 |
| YB110pSB229 | CYPIA1 + hOR | 40.8 ± 7.8 |
| GRF18pCS512 | CYP1A1 + hOR | 32.1 ± 2.5 |
| YHE2pDP34 | ND | |
| YB110pNW144 | ND | |
| GRF18pCS508 | ND |
ND, not detectable.
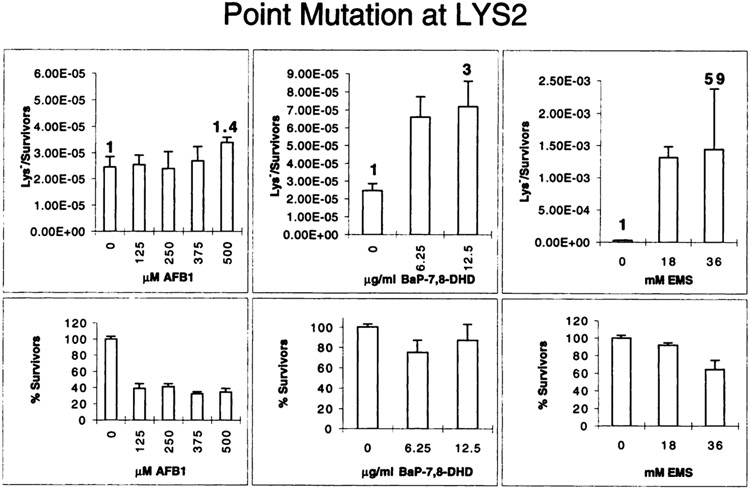
Additional evidence for functional expression of CYP1A1 in strain GRF18pCS512 was obtained from an experiment in which the strain was exposed to BaP-7,8-DHD. This strain responded to the chemical with a maximal 12-fold increase in the mutation frequency (Fig. 4H) at a survival rate of 70% (Fig. 4L), thus exhibiting an intermediate-fold increase falling between the 5-fold increase of strain YHE2pSB229 (Fig. 4B) and the 29-fold increase of strain YB110pSB229 (Fig. 4E). In conjunction with the enzyme activity measurements, this result clearly demonstrated that the observed low mutagenicity of AFB1 was not related to a problem of cytochrome P450-mediated activation of the drug. Moreover, it did not result from the selection for ura3 mutants, because strain GRF18 transformed with pCS512 or pCS518 responded very well to the directly acting carcinogen EMS. Exposing either strain to different EMS doses that reduced survival of the cells up to 12% (Fig. 4M) produced up to a 347-fold increase in the mutation frequency (Fig. 4I), which was higher than the increase observed in translocation (196-fold, Fig. 4F) or gene conversion (21-fold, Fig. 4C). As was expected, EMS exerted in both diploid strains a weaker cytotoxic effect than in the haploid strain, resulting in at least a 54% survival at the highest dose (data not shown). It was, however, possible that the URA3 gene was a particularly weak target for AFB1-mutagenesis resulting in the observed low mutation frequency. Therefore, we asked whether a low AFB1-mediated mutagenicity was also observed in the unrelated target gene LYS2. GRF18pCS512 transformants were exposed to AFB1, BaP-7,8-DHD, and EMS identically to the experiment in Fig. 4, G-I, and lys2 mutants were selected upon phenotypic expression by their resistance to α-aminoadipate (see “Materials and Methods”). Fig. 5 shows that AFB1 mutated the LYS2 gene at an even lower efficiency than the URA3 gene, resulting in a 1.4-fold increase above the spontaneous mutation frequency at 500 μM AFB1, a dose that reduced cell viability to 38%. In contrast, doses of 36 mm EMS or 12.5 μg/ml BaP-7,8-DHD increased the mutation frequencies by a factor of 59 and 3, respectively. Thus, it could be concluded that the low mutagenicity of AFB1 was not confined to URA3, and our data therefore suggest that the recombinogenic and not the mutagenic activity of AFB1 is the major constituent of the genotoxic activity of AFB1 in yeast, an observation with important implications for carcinogenesis that will be discussed below.
Fig. 5.
Chemically induced point mutations detected in the LYS2 gene. Strain GRF18pCS512 was exposed to the indicated doses of chemicals identically to the experiment in Fig. 4, G–I, and lys2 mutants were selected by their resistance toward α-amino-adipate. Mutation frequencies (top row) and the fraction of survivors (bottom row) are shown. The data represent mean and SDs from three experiments performed on 2 different days. The numbers above selected columns give the fold induction over the background frequency. Bars, SD.
DISCUSSION
We observed that AFB1, when activated intracellularly in the yeast S. cerevisiae by cytochrome P450 enzymes, strongly induced independent, mitotic recombination events that result in allelic gene conversion or reciprocal translocation (cross-overs). These recombination events constitute a probable subset of potential AFB1-induced genomic rearrangements that may either result from gene conversion or cross-overs. In contrast, AFB1-induced forward mutations in the URA3 gene were rarely detected and occurred at a 46-fold lower frequency than recombination-mediated chromosomal translocation and at a 6-fold lower frequency than gene conversion. Thus, the recombinogenic activity of activated AFB1 exceeds its mutagenic activity in yeast. This result is discussed in comparison to the recombinogenicity of other DNA damaging agents and its implication for AFB1 genotoxicity in higher eukaryotic organisms.
Several control experiments eliminated the possibility that the low mutagenic activity of AFB1 in yeast resulted from the inability to activate AFB1 to its mutagenic form. (a) The EROD activity, representing a CYP1A-specific model activity and determined in the mutation tester strain GRF18, was not different from the activities determined in the gene conversion tester strain YHE2 or the translocation tester strain YB110, arguing for similar levels of CYP1A1 and hOR expression in the three strains. (b) The polycyclic aromatic hydrocarbon BaP-7,8-DHD was efficiently activated to a mutagen in the respective GRF18 transformants, yielding a 12-fold increase over the spontaneous mutation frequency in URA3. Similar doses of BaP-7,8-DHD activated in YHE2 or YB110 transformants, however, induced only a moderate increase in the gene conversion or translocation frequency, being considerably lower than the frequencies induced by activated AFB1. (c) The alkylating agent EMS efficiently induced mutations in URA3, negating a potential problem related to the selection of ura3 mutants. (d) AFB1 gave rise to a cytotoxic response in the CYP1A1 expressing strain, but not in the control strain, arguing that AFB1 was able to enter the cell and that it was activated to a cytotoxic, but weakly mutagenic, metabolite by the CYP1A1 enzyme. (e) The low AFB1 mutagenicity was not restricted to the URA3 gene but was also observed with the unrelated LYS2 gene as target. Thus, AFB1 is unlike other mutagens, the mutagenicity of which exceed or are comparable to their recombinogenicity.
The molecular mechanism(s) that explains the high recombinogenicity of AFB1 in yeast is unknown. The stimulation of mitotic recombination in unirradiated diploids after matings with UV or X-irradiated haploids has suggested that recombination may be stimulated by the induction of trans-acting genes (55). Indeed, the levels of DNA damage-associated mitotic recombination that result in either allelic gene conversion (56) or translocation (43) are greater in a/α cells that express higher levels of damage-inducible genes (57). Alternatively, mitotic recombination can be directly stimulated by DNA lesions, either single-strand or double-strand breaks (58). Two DNA adducts induced by AFB1 include 8-hydroxydeoxyguanosine (59) and trans-8,9-dihydro-(N7-guanyl)-9-hydroxy-aflatoxin B1 (12). 8-Hy-droxydeoxyguanosine is also induced by γ-irradiation and reactive oxygen species (60), and is a substrate of formamidopyrimidine DNA glycosylase (61), resulting in apurinic sites. Because ionizing radiation-induced DNA damage and apurinic sites are recombinogenic in yeast (62), it may be that some AFB1-induced DNA lesions are also processed into substrates for homologous recombination.
It is formally possible that the TRP5 and HIS3 genes are better targets than the URA3 gene for binding of activated AFB1. A comparison of the DNA sequences revealed two stretches of five and four contiguous G residues in TRP5 and a stretch of four G residues in HIS3. As has been described for the HPRT gene (20), a DNA sequence consisting of six contiguous Gs acted as a hot spot for AFB1 binding. In contrast, the URA3 gene does not contain more than three contiguous Gs in a row, and from the study of Muench et al. (63), it is known that the second G in a contiguous trinucleotide sequence is a favored AFB1 binding site. Mutations in several of such G residues in the URA3 gene may give rise to nonconservative amino acid changes that most likely inactivate gene function. Thus, potential AFB1 binding sites are present in URA3, yet they seem not to be efficiently used for AFB1-induced mutagenesis. However, solely based on this sequence analysis, it cannot be concluded that the observed G clusters in TRP5 and HIS3 act as hot spots, and further information on the potential effect of the G-rich sequences will have to await future experiments.
Observations concerning the low mutagenicity of AFB1 in prokaryotes may provide clues for the low mutagenicity of AFB1 in yeast. Activated AFB1 is only marginally mutagenic in the Salmonella typhimurium strains TA1538 and TA1535 that carry a frameshift or a missense mutation in hisD and hisG, respectively (14). However, in the presence of plasmid pKM101, which harbors an error-prone DNA repair system and DNA polymerase, AFB1-induced frameshift and missense mutations were identified in the respective S. typhimurium strains TA98 and TA100. Thus, AFB1-mediated mutagenesis requires the induction of the SOS system (15). By analogy, the ability of AFB1 adducts to generate mutations in yeast may also arise from a secondary process or an error-prone polymerase. We speculate that the AFB1-induced DNA lesions are poor substrates for the mutagenic repair pathway, and the persistence of these lesions stimulates homologous recombination.
A relatively weak ability of activated AFB1 to induce mutations in the HPRT gene has also been observed with higher eukaryotic cell systems. The metabolically competent Chinese hamster cell line SD1 expressing CYP2B1 activated AFB1 to a metabolite that resulted in a 4.4-fold increase in the HPRT mutation frequency (64), and the AHH-1 cell line transfected to express human CYP1A2 and treated with an AFB1 dose that resulted in 25% survival also increased HPRT mutation frequency by a factor of 5 (65). Somewhat higher inductions were obtained with human lymphoblasts expressing CYP3A4 (66) or CYP1A1 (20), in which 15-fold increases in HPRT mutation frequencies were obtained at 16–20% survival. Thus, also in these higher eukaryotic cell systems, AFB1 appears not to be a strong mutagen, an observation comparable to ours with yeast.
The genetic instability induced by AFB1 in yeast may elucidate mechanisms in the progression of AFB1-induced transformation of mammalian cells. Carcinogenesis is correlated with alterations in proto-oncogenes and tumor suppressor genes (67), genes involved in either cell cycle regulation or progression. For example, mutations in p53 are found in >50% of all solid tumors (reviewed in Ref. 28). The recessivity of mutations in tumor suppressor genes implies that both copies of the wild-type tumor suppressor gene must be inactivated before gene function is lost. The rate of mutations in a given tumor suppressor gene is elevated by specific environmental carcinogens, and exposure to AFB1 is also strongly correlated with the appearance of p53 mutations detected in human HCCs. In addition, a high proportion of AFB1-related HCCs exhibit LOH at the p53 locus and other loci. For example, from 21 HCCs sampled from patients living in an AFB1-contaminated area in China (26) that also exhibited the AFB1-specific codon 249 mutation in p53, 13 HCCs had lost the second p53 allele. In a different study, LOH in the p53, Rb, and APC tumor suppressor genes was detected at a considerably higher proportion in AFB1-derived HCCs (48 of 107) compared with AFB1-unrelated HCCs (5 of 36; Ref. 27). Because the presence of two independent point mutations in both copies of a tumor suppressor gene is rarely observed, LOH is the predominate mechanism for inactivating the second allele (25, 68). Thus, the introduction of a point mutation in one tumor suppressor gene followed by LOH resulting in loss of the second allele may be a prominent mechanism of liver carcinogenesis.
Molecular mechanisms of LOH in somatic cells include chromosomal deletions, chromosomal nondisjunctions, gene conversion events, or mitotic crossing-over involving homologous chromosomes. Indeed, a single mitotic crossing-over occurring in G2 phase of the cell cycle in the target region present between the tumor suppressor locus and its corresponding centromere will lead to segregation of the tumor suppressor locus and all loci distal from the crossing-over site in half of the mitotic divisions (69). Double pericentric cross-overs between homologues thus may result in a recombined chromosome that cannot be distinguished from one that resulted from a nondisjunction event followed by reduplication.
Although homologous recombination is an attractive mechanism of LOH, mitotic recombination between homologues is repressed in normal mammalian somatic cells (70, 71). However, ataxia telangiectasia cell lines that are defective in the p53 signal transduction pathway (72) exhibit higher levels of intrachromosomal homologous recombination between direct repeated sequences. Because AFB1 promotes both sister-chromatid exchanges (33) and intrachromosomal recombination in mammalian cells (34), and because deletions of p53 have also been observed in vivo in AFB1-induced rat liver tumors (73), it is plausible that AFB1 also induces some of the genetic changes present in liver tumors by homologous recombination resulting in mitotic gene conversion or crossing-over.
In conclusion, our data argue for another genotoxic activity of metabolically activated AFB1 apart from its mutagenic activity. In the lower eukaryote S. cerevisiae, AFB1 stimulated homologous recombination that resulted in both allelic gene conversion and translocation. This recombinogenic activity of the mycotoxin by far exceeds its mutagenic activity. It is interesting to speculate that AFB1 in mammalian cells also stimulates mitotic recombination that results in allelic gene conversion and mitotic crossing-over. The future identification of specific AFB1 lesions that stimulate homologous recombination either in situ or by the activation of trans-activating proteins may provide insights into the lesions that provoke genetic instability in mammalian cells. We suggest that AFB1-associated recombination may result in LOH and efficiently contributes to liver carcinogenesis.
ACKNOWLEDGMENTS
We thank Joseph Koudelik for competent technical assistance.
This work was supported by Research Grant 0-10-253-91 (to C. S.) from the Swiss Federal Institute of Technology and Grants CA 70105 (to M. F.) from the National Cancer Institute and Leukemia Research Foundation (to M. F.).
Footnotes
The abbreviations used are: AFBI, aflatoxin B1; LOH, loss of heterozygosity; Ade, adenine; BaP-7,8-DHD, benzo[a]pyrene-7,8-dihydrodiol; EMS, ethyl methanesulfonate; EROD, 7-ethoxy-resorufin-O-deethylase; FOA, 5-fluoro-orotic acid; HCC, hepatocellular carcinoma; hOR, human NADPH-cytochrome P450-oxidoreductase; Ura, uracil; Val, valine; CHEF, contour-clamped homogeneous electric field; YM, yeast minimal.
Obtained from A. Hinnen.
Contributor Information
Christian Sengstag, Genetics Department, Institute of Toxicology, Swiss Federal Institute of Technology and University of Zürich, CH-8603 Schwerzenbach, Switzerland.
Béatrice Weibel, Genetics Department, Institute of Toxicology, Swiss Federal Institute of Technology and University of Zürich, CH-8603 Schwerzenbach, Switzerland.
Michael Fasullo, Department of Radiotherapy, Loyola University Chicago, Stritch School of Medicine, Maywood, Illinois 60153.
REFERENCES
- 1.Eaton DL, and Gallagher EP Mechanisms of aflatoxin carcinogenesis. Annu. Rev. Pharmacol. Toxicol, 34: 135–172, 1994. [DOI] [PubMed] [Google Scholar]
- 2.Groopman JD, Cain LG, and Kensler TW Aflatoxin exposure in human populations: measurements and relationship to cancer. Crit. Rev. Toxicol, 19: 113–145, 1988. [DOI] [PubMed] [Google Scholar]
- 3.IARC Mycotoxins. IARC. Monographs on the Evaluation of Carcinogenic Risks to Humans. Some Naturally Occurring Substances: Food Items and Constituents. Heterocyclic Aromatic Amines and Mycotoxins, Vol. 56, pp. 245–395. Lyon, France: IARC WHO, 1993. [Google Scholar]
- 4.Sengstag C, Eugster H-P, and Wtirgler FE High promutagen activating capacity of yeast microsomes containing human cytochrome P-450 1A and human NADPH-cytochrome P-450 reductase. Carcinogenesis (Lond.), 15: 837–843, 1994. [DOI] [PubMed] [Google Scholar]
- 5.Shimada T, Nakamura S, Imaoka S, and Funae Y Genotoxic and mutagenic activation of aflatoxin B1 by constitutive forms of cytochrome P-450 in rat liver microsomes. Toxicol. Appl. Pharmacol, 91: 13–21, 1987. [DOI] [PubMed] [Google Scholar]
- 6.Penman BW, Chen L, Gelboin HV, Gonzalez FJ, and Crespi CL Development of a human lymphoblastoid cell line constitutively expressing human CYP1A1 cDNA: substrate specificity with model substrates and promutagens. Carcinogenesis (Lond.), 15: 1931–1937, 1994. [DOI] [PubMed] [Google Scholar]
- 7.Crespi CL, Langenbach R, Rudo K, Chen YT, and Davies RL Transfection of a human cytochrome P-450 gene into the human lymphoblastoid cell line, AHH-1, and use of the recombinant cell line in gene mutation assays. Carcinogenesis (Lond.), 10: 295–301, 1989. [DOI] [PubMed] [Google Scholar]
- 8.Sawada M, Kitamura R, and Kamataki T Stable expression of monkey cytochrome P-450IA1 cDNA in Chinese hamster CHL cells and its application for detection of mutagenicity of aflatoxin B1. Mutat. Res, 265: 23–29, 1992. [DOI] [PubMed] [Google Scholar]
- 9.Sawada M, Kitamura R, Ohgiya S, and Kamataki T Stable expression of mouse NADPH-cytochrome P450 reductase and monkey P4501A1 cDNAs in Chinese hamster cells: establishment of cell lines highly sensitive to aflatoxin B. Arch. Biochem. Biophys, 300: 164–168, 1993. [DOI] [PubMed] [Google Scholar]
- 10.Monroe DH, and Eaton DL Effects of modulation of hepatic glutathione on biotransformation and covalent binding of aflatoxin B1 to DNA in the mouse. Toxicol. Appl. Pharmacol, 94: 118–127, 1988. [DOI] [PubMed] [Google Scholar]
- 11.McGlynn KA, Rosvold EA, Lustbader ED, Hu Y, Clapper ML, Zhou T, Wild CP, Xia XL, Baffoe-Bonnie A, Ofori-Adjei D, Chen G-C, London WT, Shen FM, and Buetow KH Susceptibility to hepatocellular carcinoma is associated with genetic variation in the enzymatic detoxification of aflatoxin Bl. Proc. Natl. Acad. Sci. USA, 92: 2384–2387, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin CN, and Gamer RC Aflatoxin B-oxide generated chemical or enzymatic oxidation of aflatoxin B1 causes guanine substitution in nucleic acids. Nature (Lond.), 267: 863–865, 1977. [DOI] [PubMed] [Google Scholar]
- 13.Wong J, and Hsieh DPH Mutagenicity of aflatoxins related to their metabolism and carcinogenic potential. Proc. Natl. Acad. Sci. USA, 73: 2241–2244, 1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCann J, Spingam NE, Kobori J, and Ames BN Detection of carcinogens as mutagens: bacterial tester strain with R factor plasmids. Proc. Natl. Acad. Sci. USA, 72: 979–983, 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster PL, Eisenstadt E, and Miller JH Base substitution mutations induced by metabolically activated aflatoxin Bl. Proc. Natl. Acad. Sci. USA, 80: 2695–2698, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch WH, Henrikson EN, Kupchella E, and Cebula TA Salmonella typhimurium strain TA100 differentiates several classes of carcinogens and mutagens by base substitution specificity. Carcinogenesis (Lond.), 15: 79–88, 1994. [DOI] [PubMed] [Google Scholar]
- 17.Bailey EA, Iyer RS, Stone MP, Harris TM, and Essigmann JM Mutational properties of the primary aflatoxin B1-DNA adduct. Proc. Natl. Acad. Sci. USA, 93: 1535–1539, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trottier Y, Waithe WI, and Anderson A Kinds of mutations induced by aflatoxin B1 in a shuttle vector replicating in human cells transiently expressing cytochrome P4501A2 cDNA. Mol. Carcinog, 6: 140–147, 1992. [DOI] [PubMed] [Google Scholar]
- 19.Courtemanche C, and Anderson A Shuttle-vector mutagenesis by aflatoxin B1 in human cells: effects of sequence context on the supF mutational spectrum. Mutat. Res, 306: 143–151, 1994. [DOI] [PubMed] [Google Scholar]
- 20.Cariello NF, Cui L, and Skopek TR In vitro mutational spectrum of aflatoxin B1 in the human hypoxanthine guanine phosphoribosyltransferase gene. Cancer Res., 54: 4436–4441, 1994. [PubMed] [Google Scholar]
- 21.Aguilar F, Harris CC, Sun T, Hollstein M, and Cerutti P Geographic variation of p53 mutational profile in nonmalignant human liver. Science (Washington DC), 264: 1317–1319, 1994. [DOI] [PubMed] [Google Scholar]
- 22.Li D, Cao Y, He L, Wang NJ, and Gu JR Aberrations of p53 gene in human hepatocellular carcinoma from China. Carcinogenesis (Lond.), 14: 169–173, 1993. [DOI] [PubMed] [Google Scholar]
- 23.Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, and Harris CC Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature (Lond.), 350: 427–428, 1991. [DOI] [PubMed] [Google Scholar]
- 24.Bressac B, Kew M, Wands J, and Ozturk M Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature (Lond.), 350: 429–431, 1991. [DOI] [PubMed] [Google Scholar]
- 25.Oda T, Tsuda H, Scarpa A, Sakamoto M, and Hirohashi S p53 gene mutation spectrum in hepatocellular carcinoma. Cancer Res., 52: 6358–6364, 1992. [PubMed] [Google Scholar]
- 26.Scorsone KA, Zhou YZ, Butel JS, and Slagle BL p53 mutations cluster at codon 249 in hepatitis B virus-positive hepatocellular carcinomas from China. Cancer Res., 52: 1635–1638, 1992. [PubMed] [Google Scholar]
- 27.Fujimoto Y, Hampton LL, Wirth PJ, Wang NJ, Xie JP, and Thorgeirsson SS Alterations of tumor suppressor genes and allelic losses in human hepatocellular carcinomas in China. Cancer Res., 54: 281–285, 1994. [PubMed] [Google Scholar]
- 28.Greenblatt MS, Bennett WP, Hollstein M, and Harris CC Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res., 54: 4855–4878, 1994. [PubMed] [Google Scholar]
- 29.Aguilar F, Hussain SP, and Cerutti P Aflatoxin B1 induces the transversions of G to T in codon 249 of the p53 tumor suppressor gene in human hepatocytes. Proc. Natl. Acad. Sci. USA, 90: 8586–8590, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CJ, Butel JS, and Bradley A Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature (Lond.), 356: 215–221, 1992. [DOI] [PubMed] [Google Scholar]
- 31.Tlsty TD, Margolin BH, and Lum K Differences in the rates of gene amplification in nontumorigenic and tumorigenic cell lines as measured by Luriα-Delbruck fluctuation analysis. Proc. Natl. Acad. Sci. USA, 86: 9441–9445, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin Y, Tainsky MA, Bischoff FZ, Strong LC, and Wahl GM Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell, 70: 937–948, 1992. [DOI] [PubMed] [Google Scholar]
- 33.Kulka U, Doehmer J, Glatt HR, and Bauchinger M Cytogenetic effects of promutagens in genetically engineered V79 Chinese hamster cells expressing cyto-chromes P450. Eur. J. Pharmacol, 228: 299–304, 1993. [DOI] [PubMed] [Google Scholar]
- 34.Zhang LH, and Jenssen D Studies on intrachromosomal recombination in SP5/V79 Chinese hamster cells upon exposure to different agents related to carcinogenesis. Carcinogenesis (Lond.), 15: 2303–2310, 1994. [DOI] [PubMed] [Google Scholar]
- 35.Chu G, Vollrath D, and Davis RW Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science (Washington DC), 234: 1582–1585, 1986. [DOI] [PubMed] [Google Scholar]
- 36.Zimmermann FK, Kem R, and Rasenberger H A yeast strain for simultaneous detection of induced mitotic crossing over, mitotic gene conversion and reverse mutation. Mutat. Res, 28: 381–388, 1975. [Google Scholar]
- 37.Fasullo MT, and Davis RW Direction of chromosome rearrangements in Saccharomyces cerevisiae by use of his3 recombinational substrates. Mol. Cell Biol, 8: 4370–4380, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiestl RH, Gietz RD, Mehta RD, and Hastings PJ Carcinogens induce intrachromosomal recombination in yeast. Carcinogenesis (Lond.), 10: 1445–1455, 1989. [DOI] [PubMed] [Google Scholar]
- 39.Fasullo M, Dave P, and Rothstein R DNA-damaging agents stimulate the formation of directed reciprocal translocations in Saccharomyces cerevisiae. Mutat. Res, 314: 121–133, 1994. [DOI] [PubMed] [Google Scholar]
- 40.Zimmermann FK, von Borstel RC, von Halle ES, Parry JM, Siebert D, Zetterberg G, Barale R, and Loprieno N Testing of chemicals for genetic activity with Saccharomyces cerevisiae: a report of the U. S. Environmental Protection Agency Gene-Tox Program. Mutat. Res, 133: 199–244, 1984. [DOI] [PubMed] [Google Scholar]
- 41.Sengstag C, and Würgler FE DNA recombination induced by aflatoxin B-1 activated by cytochrome P450 1A enzymes. Mol. Carcinog, 11: 227–235, 1994. [DOI] [PubMed] [Google Scholar]
- 42.Eugster HP, Sengstag C, Meyer UA, Hinnen A, and Würgler FE Consti-tutive and inducible expression of human cytochrome P450IA1 in yeast Saccharomyces cerevisiae: an alternative enzyme source for in vitro studies. Biochem. Biophys. Res. Commun, 172: 737–744, 1990. [DOI] [PubMed] [Google Scholar]
- 43.Fasullo M, and Dave P Mating type regulates the radiation-associated stimulation of reciprocal translocation events in Saccharomyces cerevisiae. Mol. Gen. Genet, 243: 63–70, 1994. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch EF, and Maniatis T Molecular Cloning: A Laboratory Manual, 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1989. [Google Scholar]
- 45.Eugster HP, Bärtsch S, Würgler FE, and Sengstag C Functional co-expression of human oxidoreductase and cytochrome P450 1A1 in Saccharomyces cerevisiae results in increased EROD activity. Biochem. Biophys. Res. Commun, 185: 641–647, 1992. [DOI] [PubMed] [Google Scholar]
- 46.Meyhack B, Hinnen A, and Heim J Heterologous gene expression in Saccharomyces cerevisiae. In: Hershberger CL, Queener SW, and Hegeman G (eds.), Genetics and Molecular Biology of Industrial Microorganisms, pp. 311–321. Washington, DC: American Society for Microbiology, 1989. [Google Scholar]
- 47.Wittekindt NE, Wiirgler FE, and Sengstag C Functional expression of fused enzymes between human cytochrome P4501A1 and human NADPH-cytochrome P450 oxidoreductase in Saccharomyces cerevisiae. DNA Cell Biol., 14: 273–283, 1995. [DOI] [PubMed] [Google Scholar]
- 48.Rose AB, and Broach JR Propagation and expression of cloned genes in yeast: 2-microns circle-based vectors. Methods Enzymol., 185: 234–279, 1990. [DOI] [PubMed] [Google Scholar]
- 49.Yanisch-Perron C, Vieira J, and Messing J Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene (Amst.), 33: 103–119, 1985. [DOI] [PubMed] [Google Scholar]
- 50.Klebe RJ, Harriss JV, Sharp ZD, and Douglas MG A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene (Amst.), 25: 333–341, 1983. [DOI] [PubMed] [Google Scholar]
- 51.Rutten AAJJL, Falke HE, Catsburg JF, Wortelboer HM, Blaauboer BJ, Doom L, van Leeuwen FXR, Theelen R, and Rietjens IMCM Interlaboratory comparison of microsomal ethoxyresorufin and pentoxyresorufin O-dealkylation determinations: standardization of assay conditions. Arch. Toxicol, 66: 237–244, 1992. [DOI] [PubMed] [Google Scholar]
- 52.Sherman F Getting started with yeast. Methods Enzymol., 194: 3–21, 1991. [DOI] [PubMed] [Google Scholar]
- 53.Thakker DR, Yagi H, Lu AYH, Levin W, Conney AH, and Jerina DM Metabolism of benzo[a]pyrene: conversion of (± )-trans-7,8-dihydroxy-7,8-dihydro-benzo[a]pyrene to highly mutagenic 7,8-diol-9,10-epoxides. Proc. Natl. Acad. Sci. USA, 73: 3381–3385, 1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimada T, Martin MV, Pruess SD, Mamett LJ, and Guengerich FP Roles of individual human cytochrome P-450 enzymes in the bioactivation of benzo(a)pyrene, 7,8-dihydroxy-7,8-dihydrobenzo(a)pyrene, and other dihydrodiol derivatives of polycyclic aromatic hydrocarbons. Cancer Res., 49: 6304–6312, 1989. [PubMed] [Google Scholar]
- 55.Fabre F, and Henschel R Genetic evidence for inducibility of recombination competence in yeast. Proc. Natl. Acad. Sci. USA, 74: 1667–1671, 1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heude M, and Fabre F a/alphα-control of DNA repair in the yeast Saccharomyces cerevisiae: genetic and physiological aspects. Genetics, 133: 489–498, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cole GM, Schild D, Lovett ST, and Mortimer RK Regulation of RAD54- and RAD52-lacZ gene fusions in Saccharomyces cerevisiae in response to DNA damage. Mol. Cell. Biol, 7: 1078–1084, 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roeder GS, and Stewart SE Mitotic recombination in yeast. Trends Genet., 4: 263–267, 1988. [DOI] [PubMed] [Google Scholar]
- 59.Shen HM, Ong CN, Lee BL, and Shi CY Aflatoxin Bl-induced 8-hy-droxydeoxyguanosine formation in rat hepatic DNA. Carcinogenesis (Lond.), 16: 419–422, 1995. [DOI] [PubMed] [Google Scholar]
- 60.Teoule R Radiation-induced DNA damage and its repair. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med, 51: 573–589, 1987. [DOI] [PubMed] [Google Scholar]
- 61.Chung MH, Kim HS, Ohtsuka E, Kasai H, Yamamoto F, and Nishimura S An endonuclease activity in human polymorphonuclear neutrophils that removes 8-hydroxyguanine residues from DNA. Biochem. Biophys. Res. Commun, 178: 1472–1478, 1991. [DOI] [PubMed] [Google Scholar]
- 62.Friedberg E, Walker GC, and Siede W DNA repair and mutagenesis in eukaryotic cells, pp. 523–576. Washington, DC: American Society for Microbiology Press, 1995. [Google Scholar]
- 63.Muench KF, Misra RP, and Humayun MZ Sequence specificity in aflatoxin B1-DNA interactions. Proc. Natl. Acad. Sci. USA, 80: 6–10, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doehmer J, Dogra S, Friedberg T, Monier S, Adesnik M, Glatt H, and Oesch F Stable expression of rat cytochrome P-450IIB1 cDNA in Chinese hamster cells (V79) and metabolic activation of aflatoxin Bl. Proc. Natl. Acad. Sci. USA, 85: 5769–5773, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crespi CL, Steimel DT, Aoyama T, Gelboin HV, and Gonzalez FJ Stable expression of human cytochrome P450IA2 cDNA in a human lymphoblastoid cell line: role of the enzyme in the metabolic activation of aflatoxin B1. Mol. Carcinog, 3: 5–8, 1990. [DOI] [PubMed] [Google Scholar]
- 66.Crespi CL, Penman BW, Steimel DT, Gelboin HV, and Gonzalez FJ The development of a human cell line stably expressing human CYP3A4: role in the metabolic activation of aflatoxin B1 and comparison to CYP1A2 and CYP2A3. Carcinogenesis (Lond.). 12: 355–359, 1991. [DOI] [PubMed] [Google Scholar]
- 67.Fearon ER, and Vogelstein B A genetic model for colorectal tumorigenesis. Cell, 61: 759–767, 1990. [DOI] [PubMed] [Google Scholar]
- 68.Murakami Y, Hayashi K, Hirohashi S, and Sekiya T Aberrations of the tumor suppressor p53 and retinoblastoma genes in human hepatocellular carcinomas. Cancer Res., 51: 5520–5525, 1991. [PubMed] [Google Scholar]
- 69.Sengstag C The role of mitotic recombination in carcinogenesis. Crit. Rev. Toxicol, 24: 323–353, 1994. [DOI] [PubMed] [Google Scholar]
- 70.Godwin AR, Bollag RJ, Christie DM, and Liskay RM Spontaneous and restriction enzyme-induced chromosomal recombination in mammalian cells. Proc. Natl. Acad. Sci. USA, 91: 12554–12558, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shulman MJ, Collins C, Connor A, Read LR, and Baker MD Interchro-mosomal recombination is suppressed in mammalian somatic cells. EMBO J., 14: 4102–4107, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meyn MS High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science (Washington DC), 260: 1327–1330, 1993. [DOI] [PubMed] [Google Scholar]
- 73.Lilleberg SL, Cabonce MA, Raju NR, Wagner LM, and Kier LD Alterations in the structural gene and the expression of p53 in rat liver tumors induced by aflatoxin Bl. Mol. Carcinog, 6: 159–172, 1992. [DOI] [PubMed] [Google Scholar]