Figure 4: Subcellular localization prediction and residues important for localization of EPIC.
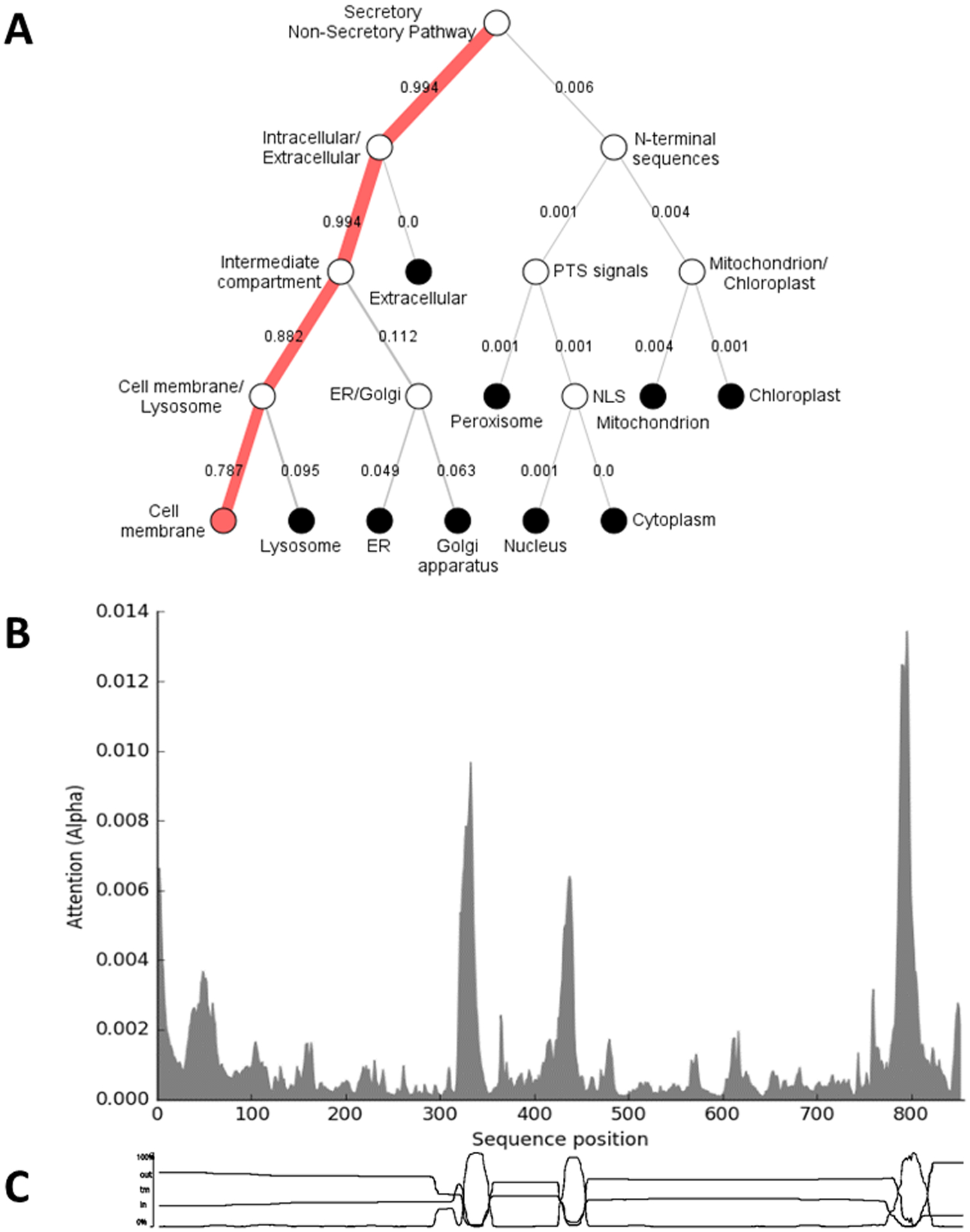
Results of the submission of the multi-epitope antigen sequence (EPIC) to the DeepLoc 1.0 Server (https://services.healthtech.dtu.dk/service.php?DeepLoc-1.0) for prediction of subcellular localization. (A) Hierarchical tree output showing the most likely localization of the antigen with numbers representing percent likelihood. (B) Graph of the importance of each residue for localization. (C) Location of the three transmembrane domains with EPIC, as shown by the transmembrane prediction function of Geneious Prime software.
