Fig. 1. MitoKO DdCBE library design strategy.
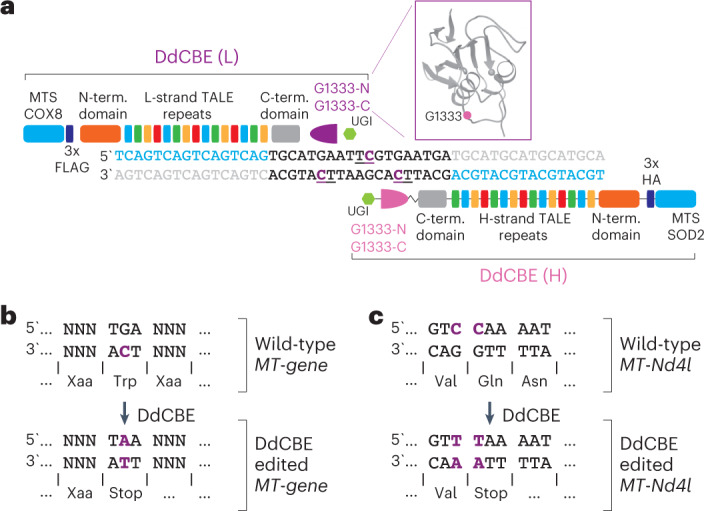
a, The architecture of DdCBE monomers used to generate the mitoKO library targeting each protein-coding gene of the mouse mtDNA. The mtDNA specificity is provided by programmable TALE domains. In each experiment, the DddAtox G1333 split (purple) was tested in both orientations to achieve editing of the desired ‘TC’ sites. The MTS were from human superoxide dismutase 2 (SOD2) or cytochrome C oxidase subunit 8 (COX8). UGI, uracil glycosylase inhibitor. L-strand or (L), light mtDNA strand. H-strand or (H), heavy mtDNA strand. b, The strategy employed to ablate the 12 out of 13 mtDNA protein-encoded genes (Nd1, Nd2, Nd3, Nd4, Nd5, Nd6, Cytb, CoI, CoII, CoIII, Atp6 and Atp8) by introducing a premature stop codon with base editing. In the vertebrate mitochondrial genetic code, the TGA codon encodes tryptophan (Trp). Transition of the cytosine (C) in the opposite strand to a thymine (T) (in purple) using base editing leads to a premature TAA stop codon. c, The strategy employed to knock out the mtDNA protein-encoded gene Nd4l, by introducing a premature stop codon with base editing. Transition of the cytosine (C) in a CAA codon encoding glutamine (Gln) to a thymine (T) (in purple) using base editing leads to a premature TAA stop codon, thereby silencing the Nd4l gene. In this site, base editing can potentially edit the adjacent C of a GTC codon encoding valine (Val). However, the resulting GTT codon also encodes valine leading to a silent mutation.
