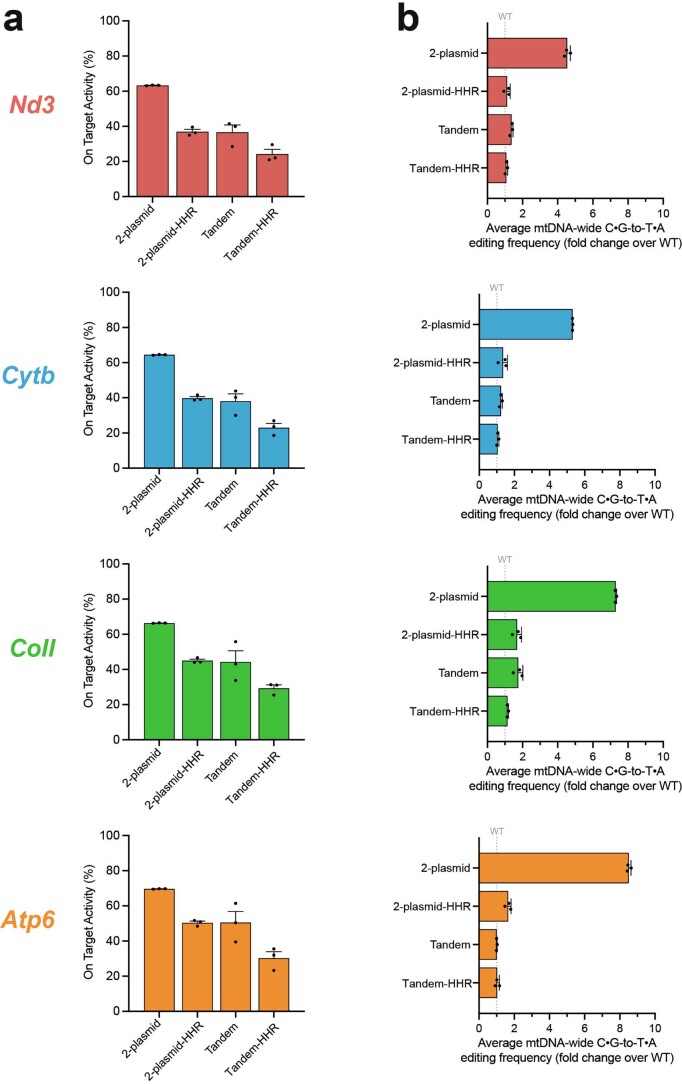
Extended Data Fig. 6. mtDNA on and off-target editing by the lead MitoKO DdCBE pairs following fine-tuned expression.
a. On-target performance of the Nd3, Cytb, CoII and Atp6 MitoKO constructs transiently delivered into NIH/3T3 cells, as separate monomers (2-plasmid), separate monomers with the hammerhead ribozyme (HHR) incorporated in mRNA (2-plasmid-HHR), bi-cistronic construct, with the tandemly arrayed DdCBE monomers linked by the T2A element (Tandem) and the tandem, T2A-linked monomers harboring HHR (Tandem-HHR). Bars represent the mean and error bars represent ± SEM (n = 3, biological replicates). Source data are provided as a Source Data file. b. Mitochondrial genome-wide off-targeting the Nd3, Cytb, CoII and Atp6 MitoKO constructs measured by NGS and presented fold change over wild-type (WT) controls. Bars represent the mean and error bars represent ± SEM (n = 3, biological replicates). Source data are provided as a Source Data file.