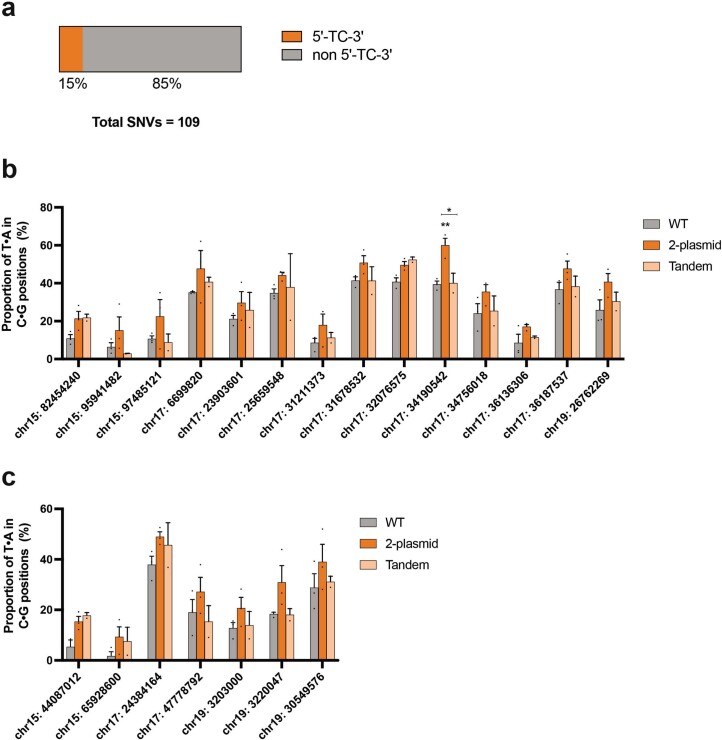
Extended Data Fig. 7. Nuclear DNA off-target analysis by the Atp6 MitoKO DdCBE pair.
a. Number of SNVs identified in chromosomes 15 to 19, where T:A nucleotides are identified in C:G positions in cells transfected with the 2-plasmid version of the Atp6 MitoKO constructs, as compared to WT cells, and the proportion of SNVs found in a 5´-TC-3´context or non 5´-TC-3´ context. b,c. NGS analysis of the proportion of T:A identified in C:G loci at single positions of chromosomes 15 to 19 that are in a (b) 5´-TC-3´ or (c) 5´-TCC-3´ context. Cells were transiently transfected (7 days) with the 2-plasmid or tandem architecture of the Atp6 MitoKO constructs and compared with WT controls. Bars represent the mean and error bars represent ± SEM (n = 3 for WT and 2-plasmid, n = 2 for Tandem, biological replicates). Ordinary two-way ANOVA with Tukeys´s test: * P-value < 0.05; ** P-value < 0.01. Source data are provided as a Source Data file.