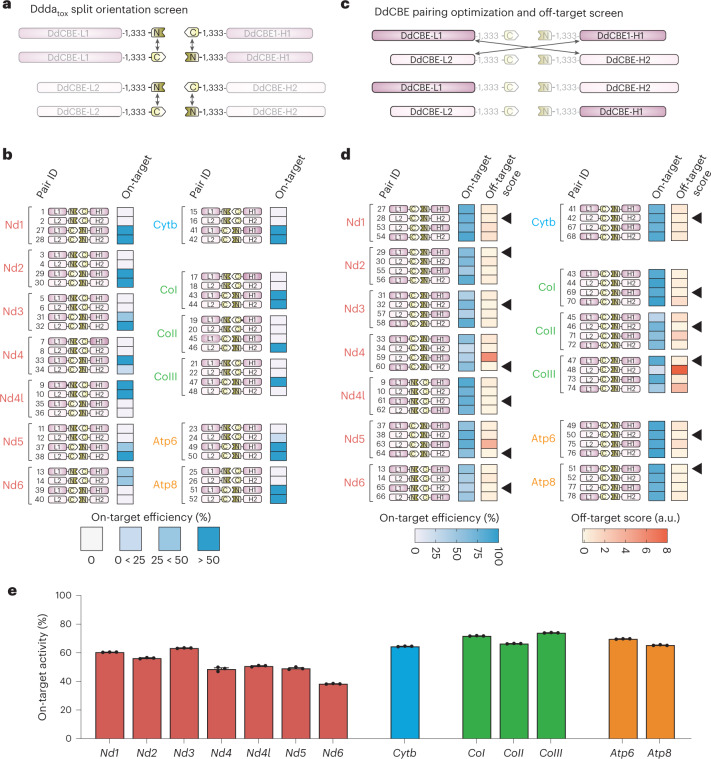
Fig. 3. Selection screening of DdCBEs for MitoKO library generation.
a, Schematic of the first screening for the optimization of DddAtox G1333 split orientation. In this screen, DdCBEs were generated by pairing TALEs L1 with H1 and L2 with H2 (Extended Data Fig. 1), testing the N-terminal fragment of DddAtox G1333 linked to L1 or L2 and the C-terminal part linked to H1 or H2, and the reciprocal orientation, with the C terminal of DddAtox G1333 linked to L1 or L2 and the N terminal linked to H1 or H2. b, In vitro on-target editing efficiency in cells after transient expression with the indicated MitoKO DdCBE pairs (Pair ID). The schematics represent the DdCBE pair combinations and which DddAtox G1333 split orientation was used. On-target efficiency was analysed by Sanger sequencing. Note the impact of split orientation on the on-target efficiency. c, Schematic of the second screening for the optimization of DdCBE pairings and off-target analysis. In this screen, DdCBEs with the most efficient DddAtox G1333 split orientation (from screen 1; b,c) were tested with additional pairings: TALEs L1 with H2 and L2 with H1. d, In vitro on-target editing efficiency and off-target scores in NIH/3T3 cells after transient expression with the indicated DdCBE pairs from the MitoKO library (Pair ID). The schematics represent the DdCBE pair combinations and which DddAtox G1333 split orientation was used. On-target efficiency and off-targets were analysed by next-generation sequencing (NGS). The final MitoKO DdCBEs (indicated with black arrows) were selected on the basis of high on-target efficiency and low off-target score (less mtDNA-wide off-targets). Source data are provided as a Source Data file. e, On-target editing by the lead MitoKO DdCBE pairs from 14 d after transfection measured by NGS. Bars and error bars represent mean ± s.e.m. (n = 3 technical replicates).