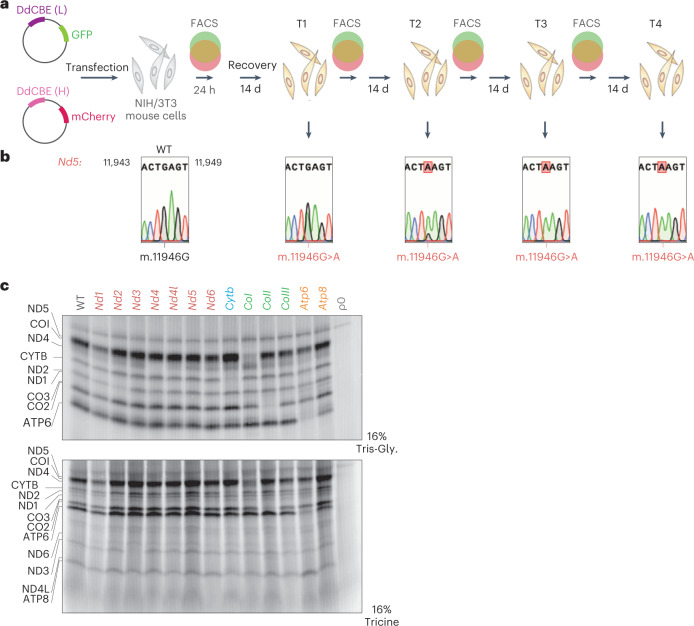
Fig. 4. Near-homoplasmic mtDNA editing using the MitoKO library.
a, Schematic of the general workflow for evaluation of the four iterative transfection and recovery cycles (T1–T4) of MitoKO pairings (for further details, see Extended Data Fig. 1c). b, Sanger sequencing of the Nd5 editing site upon iterative cycles of expression and recovery (for the remaining mtDNA genes, see Extended Data Fig. 2). c, Mitochondrial de novo protein synthesis in the NIH/3T3 cells that underwent four rounds of MitotKO DdCBE treatment, assessed by 35S-methionine metabolic labelling. WT and mtDNA-less Rho-0 cells (p0) were used as controls. The mtDNA-encoded products were resolved in two gel systems (16% Tris-Gly and 16% Tricine). For densitometric quantification, see Extended Data Fig. 3a.