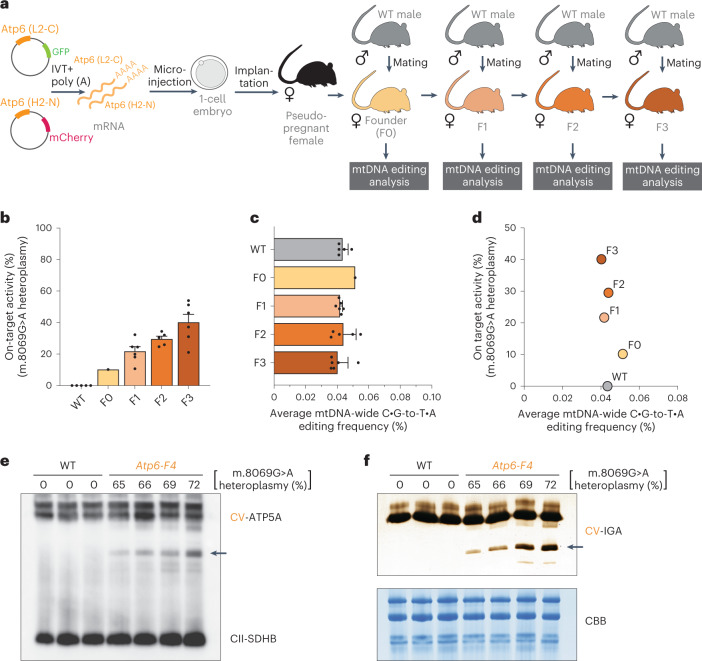
Fig. 6. In vivo mouse mtDNA editing with the MitoKO library.
a, Schematic representation of the use of Atp6 MitoKO constructs (L2-C and H2-N) for the generation of F0 founder animals carrying a heteroplasmic m.8069 G > A mutation and subsequent selective breeding scheme. IVT+poly(A), in vitro transcription and polyadenylation. b, The m.8069 G > A heteroplasmy (Y axis) in the F1–F3 generations of the selectively bred founder F0 female. Bars and error bars represent the mean ± s.e.m. c, Off-target editing in the F0–F3 m.8069 G > A animals and WT controls. Bars and error bars represent the mean ± s.e.m. d, On-target (Y axis) vs off-target (X axis) editing in the F0–F3 m.8069 G > A animals and WT controls. e, Immunoblotting of mitochondria isolated from skeletal muscle of F4 generation Atp6 m.8069 G > A heteroplasmic mice and WT controls upon a CNGE with a CV-specific antibody (ATP5A). Arrow indicates CV subcomplexes. An antibody specific for complex II (SDHB) was used as loading control. f, CNGE followed by in-gel ATP hydrolysis activity of CV in mitochondria isolated from skeletal muscle of F4 generation Atp6 m.8069 G > A heteroplasmic mice and WT controls. Arrow indicates the activity of CV subassemblies. Coomassie brilliant blue (CBB) was used as loading control. For e and f, uncropped scans are provided as a Source Data file.