Abstract
Background
Seagull as a migratory wild bird has become most popular species in southwest China since 1980s. Previously, we analyzed the gut microbiota and intestinal pathogenic bacteria configuration for this species by using 16S rRNA sequencing and culture methods. To continue in-depth research on the gut microbiome of migratory seagulls, the metagenomics, DNA virome and RNA virome were both investigated for their gut microbial communities of abundance and diversity in this study.
Results
The metagenomics results showed 99.72% of total species was bacteria, followed by viruses, fungi, archaea and eukaryota. In particular, Shigella sonnei, Escherichia albertii, Klebsiella pneumonia, Salmonella enterica and Shigella flexneri were the top distributed taxa at species level. PCoA, NMDS, and statistics indicated some drug resistant genes, such as adeL, evgS, tetA, PmrF, and evgA accumulated as time went by from November to January of the next year, and most of these genes were antibiotic efflux. DNA virome composition demonstrated that Caudovirales was the most abundance virus, followed by Cirlivirales, Geplafuvirales, Petitvirales and Piccovirales. Most of these phages corresponded to Enterobacteriaceae and Campylobacteriaceae bacterial hosts respectively. Caliciviridae, Coronaviridae and Picornaviridae were the top distributed RNA virome at family level of this migratory animal. Phylogenetic analysis indicated the sequences of contigs of Gammacoronavirus and Deltacoronavirus had highly similarity with some coronavirus references.
Conclusions
In general, the characteristics of gut microbiome of migratory seagulls were closely related to human activities, and multiomics still revealed the potential public risk to human health.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-023-09379-1.
Keywords: Gut microbiome, Migratory seagulls, DNA virome, RNA virome
Background
China is one of the countries with the most abundant animal resources in the world [1]. As the important biological resources, wild animals play the pivotal role in maintaining ecological balance and biodiversity [2]. The possibility of animal migrations to disseminate pathogens across different geographic areas has brought about several studies on the interactions between migration and infection [2–4]. Migratory birds can carry pathogens on their migrations, especially those that do not significantly affect their health. The research on infectious diseases of wild birds started relatively late in China. After the world’s first large-scale highly pathogenic avian influenza outbreak in Qinghai Lake in 2005, much more attention has been paid for investigate the migratory wild birds [5–7].
Seagull (Larus ridibundus) as a migratory wild bird has become most popular species in southwest China since 1980s. Each year, large numbers of seagulls migrate from Siberia to Southern China to overwinter, and many citizens come to feed them on parks or rivers. Thus, the public are in close contact with migratory seagulls. Previously, we analyzed the gut microbiota and intestinal pathogenic bacteria configuration for this species by using 16S rRNA sequencing and culture methods respectively [8]. Although there was little cross-infection between humans and seagulls in previous study, many pathogenic bacteria were isolated from feces of this animal, such as enteropathogenic Escherichia coli, Salmonella spp. and Yersinia spp. [8]. However, the amplicon sequencing and culture-based studies only reflect limited findings of microbial communities of animals. Now, advanced methods have been used in-depth analysis on the connection between bird activities and disease dissemination, particularly, the metagenomics and meta-transcriptomics provide us the broader characterization of microbe diversity, including bacteria, virus and fungi [9–11]. In this study, the metagenomics, DNA virome and RNA virome of migratory seagulls were both investigated for their gut microbiome of abundance and diversity.
Results
Metagenomics
The average effect reads of total samples was 99.74% ± 0.05%, and Q30 value after quality control was 89.83% ± 1.36%. 0.28% ± 0.052% of host reads was filtered. The average assembled contigs was 58,350 ± 33,306 of total samples, and predicted gene numbers was 68,462 ± 28,605.
The KEGG pathway results indicated that 103,321 genes annotated in metabolism, followed by 19,361 in environmental information processing, 10,820 in cellular processes, 8,334 in genetic information processing, 8,182 in human diseases and 3,161 in organismal systems, as Additional file 1a shown. The heatmap of top 25 pathways revealed highly relative abundance of genes distributed in HG group, especially for HG-3 and HG-4 (Additional file 1b). The eggNOG function classification showed cell wall/membrane/envelope biogenesis, transcription, amino acid transport and metabolism were the top distributed genes (Additional file 1c). The CARD annotation demonstrated that macB, bcrA, OmpK37, msbA, tetA(58), evgS, AbaF, cpxA, oleC and adeL were the top 10 relative abundance of antibiotic resistant genes. Most of these genes belonged to antibiotic efflux, as Additional file 1d shown. The top 10 distributed virulence factors between HG and HXQ groups were different (Additional file 1e). Polar-flagella, BfmRS and CdpA had highly relative abundance in HG group, compared to Capsule, AcrAB and Enterobactin in HXQ group. The PHI annotation results revealed reduce virulence (1,494), unaffected pathogenicity (732) and increased virulence (244) were the top three phenotype classifications, as Additional file 1f shown.
The composition of gut microbiome indicated that 99.72% of total species was bacteria, followed by viruses (0.2%), fungi (0.07%), archaea (0.001%) and eukaryota (0.0002%). Proteobacteria (95.7%) and Firmicutes (4%) were the most distributed abundance at phylum level of bacteria, and Gammaproteobacteria (Class level) accounted for 99.7% of Proteobacteria. The relative abundance of each sample at different taxonomic levels of bacteria was shown in Additional file 2. In general, Proteobacteria, Gammaproteobacteria, Enterobacterales, Enterobacteriaceae, Escherichia and Escherichiacoli were the most distributed abundance of each sample at different taxonomic levels, except HG-4 sample. In particular, Shigella sonnei, Escherichia albertii, Klebsiella pneumonia, Salmonella enterica and Shigella flexneri were the top distributed relative abundance at species level, and all these bacteria were considered as human pathogens (Additional file 2).
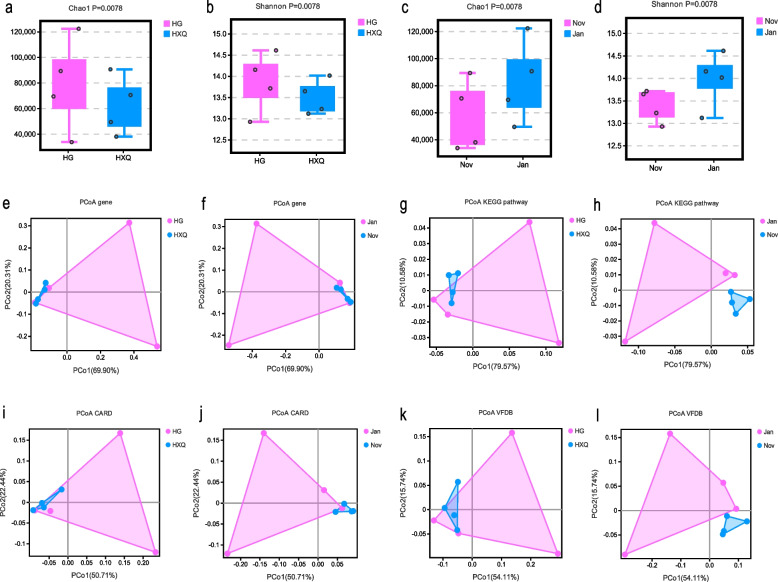
Alpha diversity showed the Chao1, Ace, Shannon and Simpson index of samples were 70,348.40 ± 28,041.63, 70,217.06 ± 28,062.91, 13.67 ± 0.54 and 0.99 ± 0.0005 respectively. Chao1 and Shannon index showed the statistical significance (P = 0.0078) between HG and HXQ groups (Fig. 1a and b), and significant differences were also found between Nov and Jan groups for two alpha diversity index (Fig. 1c and d). Beta diversity indicated that no statistical significance were found both for species composition of sample location groups (HG vs HXQ, P = 0.297) and time groups (Nov vs Jan, P = 0.356), as Fig. 1e and f shown. PCoA of KEGG pathway showed that two clusters were generated according to location and time groups respectively (Fig. 1g and h), but Anosim only revealed the statistical difference between HG and HXQ group (P = 0.028). PCoA of CARD showed the similar clustering results with KEGG pathway (Fig. 1i and j), and statistical significance was found between Nov and Jan group (P = 0.02). Anosim analysis demonstrated no differences were found both for location and time groups of VFDB diversity, although independent clustering of each group was identified (Fig. 1k and l).
Fig. 1.
Alpha and Beta diversity of metagenomic of seagulls’ feces. a The Chao1 index between HG and HXQ group. b The Shannon index between HG and HXQ group. c The Chao1 index between Nov and Jan group. d The Shannon index between Nov and Jan group. e The PCoA of annotated genes between HG and HXQ group. f The PCoA of annotated genes between Nov and Jan group. g The PCoA of KEGG pathways between HG and HXQ group. h The PCoA of KEGG pathways between Nov and Jan group. i The PCoA of annotated CARD between HG and HXQ group. j The PCoA of annotated CARD between Nov and Jan group. k The PCoA of annotated VFDB between HG and HXQ group. l The PCoA of annotated VFDB between Nov and Jan group
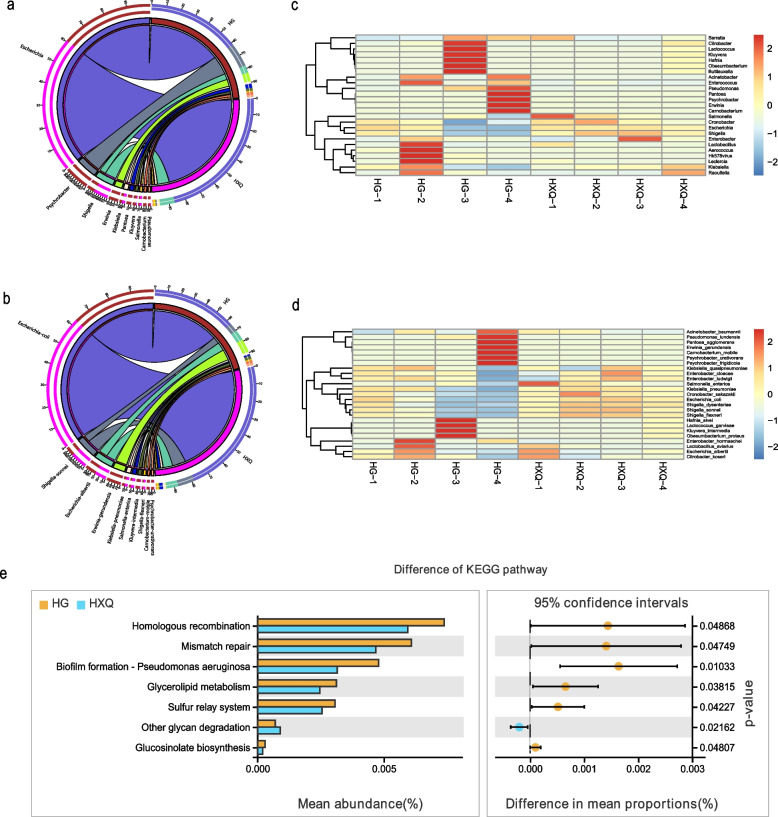
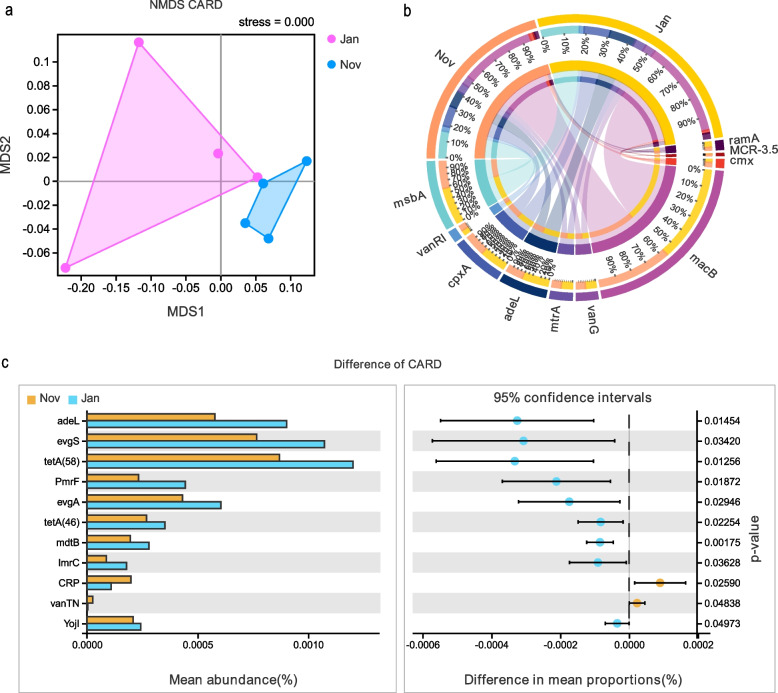
Difference analysis indicated that highly species composition was found in HG group compared to HXQ group. At genus level, Escherichia, Psychrobacter, Shigella, Erwinia, Kluyvera, Pantoea, Carnobacterium and Pseudomonas were identified in HG group (Fig. 2a). At species level, E. coli, S. sonnei, E. albertii, E. gerundensis, K. pneumonia, S. enterica, K. intermedia, C. mobile and P. urativorans had highly relative abundance in HG group in accordance with genus level (Fig. 2b). The distribution of top 25 of species composition in each group was different as well. The highly diversity was found in HG group (from HG-1 to HG-4), but S. enterica, K. pneumonia, S. dysenteriae, S. sonnei and S. flexneri had highly relative abundance in HXQ group (from HXQ-1 to HXQ-4), as Fig. 2c and d shown. Welch’s t test of KEGG pathway showed homologous recombination, mismatch repair, biofilm formation, glycerolipid metabolism and sulfur relay system had highly relative abundance in HG group (Fig. 2e). In addition, the NMDS of CARD between Nov and Jan group showed two independent clusters, and Anosim indicated the statistical significance (R = 0.333, P = 0.02), as Fig. 3a shown. The circos diagram demonstrated the top 10 relative abundance of CARD annotated genes between Nov and Jan group; macB, msbA, cpxA, adeL, mtrA and vanG genes had highly abundance in Jan group (Fig. 3b). Welch’s t test revealed adeL, evgS, tetA, PmrF, evgA, mdtB, lmrC and YojI genes had higher abundance in Jan group compared to Nov group (Fig. 3c). Most of these genes were antibiotic efflux, and resistant to fluoroquinolone, macrolide, tetracycline and peptide antibiotics.
Fig. 2.
Comparison the difference of species composition between HG and HXQ group. a The circos diagram of species composition at genus level between HG and HXQ group. b The circos diagram of species composition at species level between HG and HXQ group. c. The heatmap of distribution for top 25 of species composition in each group at genus level. d The heatmap of distribution for top 25 of species composition in each group at species level. e The statistical significance of KEGG pathway using Welch’s t test between HG and HXQ group
Fig. 3.
Beta diversity and statistics of CARD annotations between Nov and Jan group. a The NMDS of CARD annotation results between Nov and Jan group. b The circos diagram of top 10 relative abundance of annotated CARD genes between Nov and Jan group. c The statistical significance of CARD annotated genes using Welch’s t test between Nov and Jan group
DNA virome
The average effect reads of total samples was 76.03% ± 8.41% for DNA virome, and 99.89% ± 0.23% of clean reads was obtained after host reads filtration. The average assembled contigs was 64,856 ± 38,055 of total samples. Virus identification and classification results showed 78.30% (152,724/195,046) of contigs was identified as phage, 1.15% (2,248/195,046) was other virus, and 20.55% (40,074/195,046) was unassigned (Additional file 3a). The dsDNA virus accounted for 77.38% (150,930/195,046), ssDNA virus accounted for 1.72% (3,353/195,046), and 20.90% (40,761/195,046) was unassigned (Additional file 3b). UniProt functional annotation results indicated 37,436 genes annotated for technical term, 25,421 for molecular function, and 24,609 for biological process, as Additional file 3c shown. KEGG annotations at three levels revealed most of genes belonged to metabolism, genetic information processing, signaling and cellular processes, DNA replication proteins, DNA repair and recombination proteins, replication and repair (Additional file 3d to f).
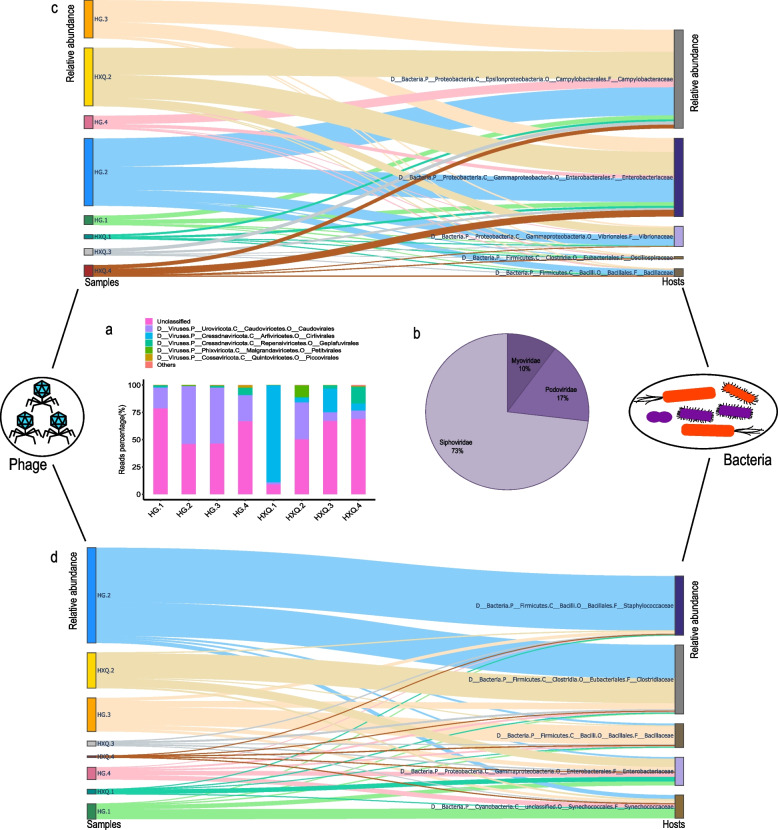
Viral composition results at order level demonstrated that Caudovirales (24.81%) was the most abundance DNA virus, followed by Cirlivirales (15.37%), Geplafuvirales (3.47%), Petitvirales (1.6%) and Piccovirales (0.41%). 54.09% of total reads was unclassified (Fig. 4a). Siphoviridae(72.52%), Podoviridae(16.80%) and Myoviridae(9.88%) constituted most of Caudovirales (Fig. 4b) at family level. Phage-host prediction based on Space Pharer showed the top 5 of relative abundance of bacterial hosts were Campylobacteraceae, Enterobacteriaceae, Vibrionaceae, Oscillospiraceae and Bacillaceae at family level corresponded to each sample (Fig. 4c). Staphylococcaceae, Clostridiaceae, Bacillaceae, Enterobacteriaceae and Synechococcaceae were the top 5 relative abundance of bacterial hosts according to VPF-Class prediction (Fig. 4d).
Fig. 4.
The characteristics of gut DNA virome of migratory seagulls. a The histogram of DNA viral composition results at order level for each sample. b The constitution of Caudovirales at family level of migratory seagulls for total samples. c The phage-host prediction based on Space Pharer. d The phage-host prediction based on VPF-Class prediction
Alpha diversity showed the Chao1, Ace, Shannon and Simpson index of samples were 15,335.93 ± 11,516.37, 15,700.62 ± 11,896.91, 8.14 ± 1.96 and 0.96 ± 0.047 respectively. Chao1 and Shannon index showed the statistical significance (P = 0.048 and 0.03) between HG and HXQ groups (Additional file 4a and b), but no significant differences found between Nov and Jan groups (Additional file 4c and d). PCoA analysis indicated two clusters were generated according to location and time groups respectively (Additional file 4e and f). However, Anosim only revealed the statistical difference between Nov and Jan group (P = 0.027).
LEfSe analysis showed the significantly different abundance between HG and HXQ group at viral order level (Additional file 4 g). The biomarker of HG group was Caudovirales compared with Cirlivirales in HXQ group. The biomarkers of different abundance between Nov and Jan group at viral order level were Geplafuvirales, Piccovirales, Kalamavirales and Rowavirales (Additional file 4 h). In addition, several contigs were also showed significant abundance between Nov and Jan group, as Additional file 4i shown. There were 17 contigs of biomarkers in Nov group compared with 34 contigs in Jan group.
RNA virome
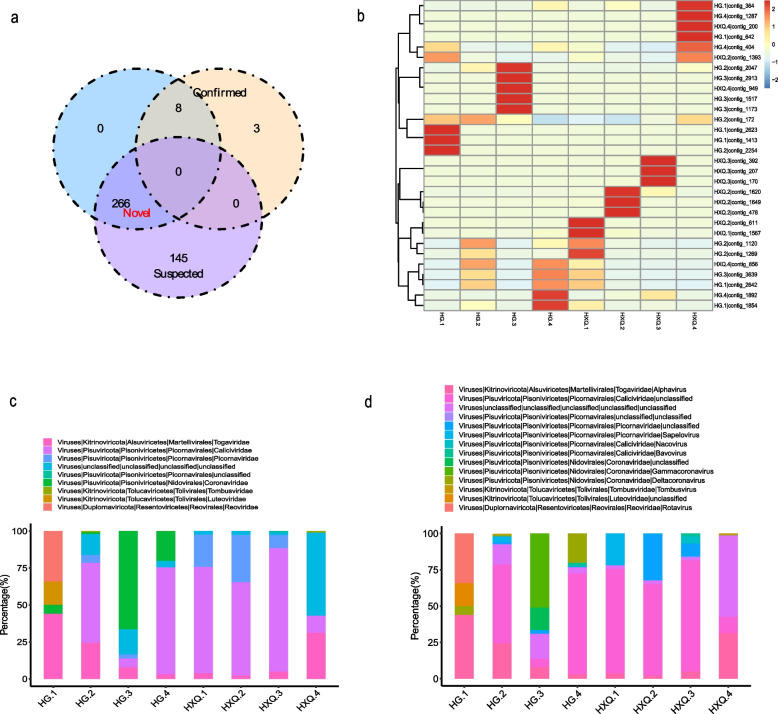
The average effect reads of total samples was 73.52% ± 1.64% for RNA virome, and 71.78% ± 8.58% of clean reads was obtained after host reads filtration. The average assembled contigs was 2,392 ± 898.03 of total samples. Based on reference virus identification method, 11 contigs of RNA viruses were recognized as confirmed, and 411 were considered as suspected. The Denovo method showed 274 contigs of RNA viruses were identified as novel. The comparison results of two methods were shown in Fig. 5a, the confirmed and novel contigs of RNA viruses were used for next step analysis. 50.54% (140/277) of contigs were annotated as Caliciviridae, followed by Coronaviridae (24.55%, 68/277) and Picornaviridae (5.42%, 15/277). The heatmap of top 30 abundance of viruses indicated that each sample had the special abundance distribution feature (Fig. 5b), and highly relative abundance of RNA viruses contigs were shown in different samples. Caliciviridae, Picornaviridae, Togaviridae, Coronaviridae, Reoviridae, Luteoviridae and Tombusviridae were the top 10 abundance of viruses at family level for RNA virome (Fig. 5c). Alphavirus, Gammacoronavirus, Deltacoronavirus, Tombusvirus, Rotavirus, Nacovirus and Bavovirus were the top 10 abundance of viruses at genus level (Fig. 5d).
Fig. 5.
The characteristics of gut RNA virome of migratory seagulls. a The venn diagram for comparison of two methods of RNA virome identification. b The heatmap of top 30 abundance of RNA viruses contigs in each sample. c The top10 abundance of viruses at family level for RNA virome. d The top 10 abundance of viruses at genus level for RNA virome
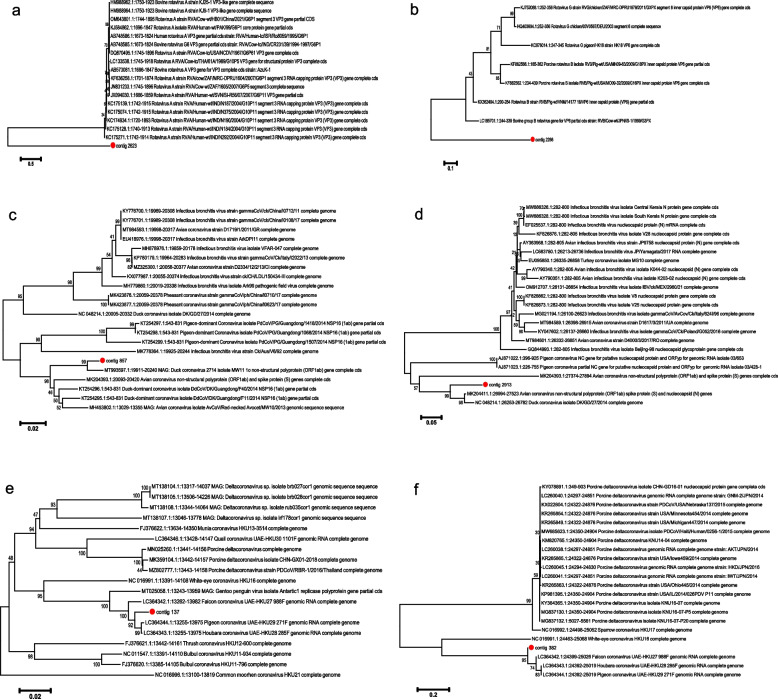
Phylogenetic analysis indicated contig 2623 belonged to Rotavirus A, showing 71.82% similarity with reference (GenBank accession: KC175271) (Fig. 6a and Additional file 5). Contig 2266 showed the 75% similarity with Rotavirus G (GenBank accession: HQ403604.1), as Fig. 6b and Additional file 5 shown. Contig 857 and contig 2913 were both belonged to Gammacoronavirus, showing 96.67% and 89.25% similarity with Duck coronavirus 2714 and Avian coronavirus respectively (GenBank accession: MT993597.1 and NC_048214.1) (Fig. 6c, d and Additional file 5). Contig 137 and 382 were Deltacoronavirus, showing 97.78% and 96.82% similarity with Houbara coronavirus UAE-HKU28 and Pigeon coronavirus UAE-HKU29 (GenBank accession: LC364343.1 and LC364344.1), as Fig. 6e, f and Additional file 5 shown.
Fig. 6.
Phylogenetic analysis of some contigs of RNA virome with references. a Phylogenetic tree of contig 2623 with Rotavirus A references. b Phylogenetic tree of contig 2266 with Rotavirus G references. c Phylogenetic tree of contig 857 with Gammacoronavirus references. d Phylogenetic tree of contig 2913 with Gammacoronavirus references. e Phylogenetic tree of contig 137 with Deltacoronavirus references. f Phylogenetic tree of contig 382 with Deltacoronavirus references
Verification of NGS data
Real-time PCR for Salmonella spp. showed positive results for all metagenomic nucleic acid samples, while 20 environmental samples (including 10 soil and 10 water samples) were negative for PCR detection (Additional file 6a). The identical results were found in Additional file 6b for Shigella spp. detection.
PCR and amplicon sequencing of contig 2623 in RNA virome showed that the NGS nucleotide sequence of contig 2623 was identical to that of the PCR amplicon by sequence alignment (Additional file 6c). In addition, NGS results for contig 857 and contig 137 in RNA virome were fully consistent with PCR amplicon sequencing results, as Additional file 6d and e shown.
Discussion
Due to their geographical dispersal ability and wide distribution range, birds not only have strong environmental adaptability, but also bring potential risks to the rapid spread of diseases [12]. Since the twentieth century, bird diseases represented by avian cholera, West Nile fever, and avian influenza have occurred frequently, resulting in the death of numerous wild birds, poultry and even humans [13, 14]. Several studies showed the gut microbiota of waterfowl, using culture-based methods and mainly concerned on some pathogens [15, 16]. Because culture-based studies only provide a limited finding of natural microbial communities, culture-independent molecular methods can be used to describe the composition of waterfowl gut microbiome. Our previous results showed Enterococcaceae, Enterobacteriaceae, and Mycoplasmataceae were the most predominant families of gut microbiota of seagulls by 16S rRNA amplicon sequencing. The most isolated intestinal pathogenic bacteria from seagulls were enteropathogenic E. coli and Salmonella by culture-based method. The results of the homology analysis of the strains by PFGE method showed that there was little cross-infection between humans and gulls, despite close proximity [8]. In this study, we used metagenomic and meta-transcriptomics techniques to in-depth reveal the features of gut microbiome of migratory seagulls. The results demonstrated that S. sonnei, E. albertii, K. pneumonia, S. enterica and S. flexneri were the top distributed taxa at species level by metagenomic investigation. All these bacteria were considered as human pathogens, and it was still a potential public risk to human health.
Birds are commonly recognized as key species in dissemination of antimicrobial agents through their feces or polluted water [12, 13, 17]. The problem is that migratory birds can fly over thousands of distances, enabling dissemination across continents. The gulls have been found to be reservoirs of beta-lactams resistance in Europe, especially for Spain [18]. In this study, some drug resistance genes accumulated as time went by from November to January of the next year. PCoA, NMDS, and statistics indicated some CARD genes had higher abundance in Jan group compared to Nov group, and most of these genes were antibiotic efflux. This phenomenon probably reflected that when seagulls migrated to Kunming city, the abundance of some drug resistant genes in their entero-intestinal tracts gradually increased. In addition, the diversity of species of HG was higher than that of HXQ. HG is closer to the wild environment, and HXQ is in the human habitation environment, which may be the reason for the difference. In KEGG pathway, the types of HG metabolic pathway genes were significantly higher than those of HXQ. As the taxonomic composition, the heatmaps of species and genus level showed the diversity of HG was significantly higher than the distribution of HXQ. The proportion of Salmonella, Shigella, Klebsiella and other related human pathogens was relatively higher in HXQ group, while highly diversity of species was shown in HG group. From another side, it was also revealed the relative restriction of living environment of seagulls once they settled down in a certain region. From the annotation of drug resistance and distribution of gut microbiome in different locations, the higher abundance of pathogenic bacteria and CARD genes were found in region closer to human living area. However, seagulls lived in wild-like environment had highly biodiversity of gut microbial community, fewer pathogenic bacteria and drug resistant genes.
Bacteriophages can affect composition of microbial communities, abundance and interact with their hosts frequently. Until now, most of the recognized phages are dsDNA, tailed phages belonging to the Caudovirales [19]. For the DNA virome of seagulls, Caudovirales was also the most abundance DNA virus. Siphoviridae, Podoviridae and Myoviridaeconstituted most of Caudovirales in this study. Coronavirus have caused three worldwide epidemics in recent two decades, including the ongoing pandemic of SARS-CoV-2 [20]. Large numbers of animals can serve as reservoirs of Coronavirus, including mice, birds and bats [21]. Alphacoronavirus and Betacoronavirus infect mammals, while Gammacoronavirus and Deltacoronavirus infect birds. Chu et al. identified three strains of Deltacoronavirus in fecal samples of migratory seagulls in Yunnan province [22]. The highly similarity of genomes and amino acid sequence to the homologs of falcon coronavirus UAE-HKU27, houbara coronavirus UAE-HKU28, and pigeon coronavirus UAE-HKU29 were both found. They concluded that there might be cross-border transmission of these avian Deltacoronavirus through specific routes. Interestingly, in this study we also found Gammacoronavirus and Deltacoronavirus of gut RNA virome in migratory seagulls. Phylogenetic analysis indicated the sequences of contigs of Deltacoronavirus had highly similarity with Houbara coronavirus UAE-HKU28 and Pigeon coronavirus UAE-HKU29 as mentioned above. Therefore, coronaviruses were possibly ubiquitous in the gut of migratory seagulls.
Conclusions
In this study, the metagenomics, DNA virome and RNA virome were both used to analyze the gut microbiome of migratory seagulls in southwest China. Some human entero-pathogens were the top distributed taxa by metagenomic investigation, and some antibiotic resistant genes were accumulated as time went by. Bacteriophages were the most abundance of DNA virome corresponded to Enterobacteriaceae and Campylobacteriaceae bacterial hosts respectively. Rotavirus, Gammacoronavirus and Deltacoronavirus were commonly found in RNA virome in this animal, and Gammacoronavirus contigs found in the present study might co-relate with the human coronavirus infection. In general, multiomics of gut microbiome for migratory seagulls still revealed the potential public risk to human health.
Methods
Sample collection
A total of 160 seagull fecal samples were collected at November 2021 and January 2022. Samples were collected according to two time points (Nov and Jan) and locations (HXQ and HG), and 80 samples were collected from one place at single time point, and the sampling from 2 selected locations was done simultaneously. HXQ is a central park in the urban area, which is closer to the living environment of the human. HG is the surrounding area of Dianchi Lake of Kunming city, which is far from the living environment of the people and closer to the natural environment. The distance between the two sampling locations was approximately 11 km. Apart from the migratory seagull, no other birds of the same species were active and defecating in the above mentioned areas. The rationale for choosing these two sites was mainly based on the proximity of one sampling site to the natural environment and the other to areas of human activity. Every 20 samples of each group were mixed into one for subsequent experimental analysis, and the mixed sample was collected around 5–10 m far from each other for each sampling location or time point.
As soon as the seagull feces were excreted, we collected and stored them in sterile containers immediately to avoid the environmental contamination. Meanwhile, 20 environmental samples included 10 soil and 10 water samples were also collected from the surrounding areas of the seagulls at the same time points.
Metagenomics
Genomic DNA was extracted using fecal sample DNA extraction kits (Tiangen, Beijing) according to the manufacturer’s instructions. The DNA quality was detected using Qubit 2.0 (Thermo Fisher Scientific) and Nanodrop accordingly. Qualified genomic DNA was fragmented by sonication to a size of 350 bp, and then end-repaired, A-tailed, and adaptor ligated using NEB Next DNA Library Prep Kit for Illumina (NEB, USA) according to the instructions. DNA fragments with length of 300–400 bp were enriched by PCR, and purified using AMPure XP system (Beckman Coulter, USA). The libraries were analyzed for size distribution by 2100 Bio-analyzer (Agilent, CA) and quantified using real-time PCR [23]. Finally, genome sequencing was performed on the Illumina Novaseq 6000 by pair-end 150 (PE150) reagent kit.
Raw data were filtered using FASTP (version 0.18.0). Clean reads of each sample were assembled individually using MEGAHIT (version 1.1.2) stepping over a k-mer range of 21 to 141 to generate sample (or group)-derived assembly [24]. Genes were predicted based on the final assembly contigs (> 500 bp) using MetaGeneMark (version 3.38) [25]. The reads were aligned to predict genes using Bowtie (version 2.2.5) to count reads numbers [26]. Final gene catalogue was obtained from non-redundant genes with gene reads count > 2. The plots were graphed using R ggplot2 package [27]. The unigenes were annotated using DIAMOND (version 0.9.24) by aligning with the deposited ones in different databases including Nr, KEGG, eggNOG, CAZy, PHI, VFDB, and CARD [28–31]. Circular layout representations of functional gene abundance were graphed using circos (version 0.69–3) [32].
Chao1, ACE, Shannon, Simpson index were calculated using Python package (version 0.5.6). Alpha index comparison between groups was calculated by Welch’s t-test and Wilcoxon rank test. Bray–curtis distance matrix based on gene/taxon/function abundance was generated by R Vegan package. Multivariate statistical techniques including PCA, PCoA and NMDS were calculated and plotted using R ggplot2 package. Adonis and Anosim test was calculated using R project Vegan package [33]. Heatmap graph were plotted using R Pheatmap package.
DNA virome
Two grams of each fecal sample were used and added 5 volumes of pre-cooled sterile SB buffer, vortex for 5 min. Centrifuged the sample at 12,000 g for 5 min to remove the precipitate, removed the cell fragments by 0.45 μm and 0.22 μm filter membrane, and the supernatant was transferred to the ultracentrifugation tube containing 28% sucrose, then centrifuged at 160,000 g for 2 h at 4 °C with HIMAC CP 100 WX ultracentrifuge (Hitachi, Japan). After removing the supernatant, resuspended the pellet in 200 μl SB buffer, added a mixture of DNase I (TaKaRa) and RNase A (TaKaRa) to digest unprotected nucleic acid and incubated at 37 °C for 60 min; added SS solution (2 μl) to inactivate the enzyme reaction at 65–75 °C for 10 min, centrifuged at 2,000 g for 5 min, and stored 200 μl of the supernatant at -20 °C [34].
DNA viruses in samples were extracted by using Magen DNA extraction kits (Magen, China), and the whole genome was amplified with Qiagen kit (REPLI-g Cell WGA & WTA Kit). The DNA quality was detected using Qubit 4.0 (Thermo Fisher Scientific) and 1.5% agarose electrophoresis. Sequencing libraries were generated using NEB Next Ultra DNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations. The library was sequenced on an Illumina Novaseq 6000 and 150 bp paired-end reads were generated.
The raw data were filtered using Soapnuke (v1.5.6) to acquire the clean data [35]. BWA software (v0.7.17) was used to compare clean reads with ribosome database (Silva.132) and host database respectively. The clean data were assembled by Megahit software (v1.1.2). The potential viral sequences in the assembled sequence were first predicted using CheckV software [36]. The virus identification was performed again using Virsorter2 software [37]. The distribution of virus reads according to the annotation results of virus contigs were counted by BWA software, and the RPKM of each virus contigs were calculated [38]. Metagenemark (v3.38) was used to predict the gene sequence of the virus contigs and evaluate the predicted gene numbers. The protein sequence of gene was compared with the virus sequence of UniProtKB/Swissprot database (ViralZone, https://viralzone.expasy.org/) by using Blastp software (v2.9.0 +), and the best hit comparison result of e < 1e-3 was screened to obtain the virus function information [39].
The abundance and diversity of DNA viruses were reflected by the analysis of Alpha and Beta Diversity as mentioned above. LDA Effect Size (LEfSe) analyse was used to find the biomarker of each group based on RPKM of virus sequence. CRISPR Recognition Tool (CRT, http://www.room220.com/crt/) was used to construct the CRISPR-CAS spacer database from bacterial genome of RefSeq database. The blastn-short (v2.9.0 +) was used to compare the identified viruses with CRISPR CAS spacer database, the best hit was selected as the possible host information of phage when the e-value < 1e-10, the comparison similarity was more than 95%, and the coverage of spacer was more than 80%.
RNA virome
The RNA viruses of enrichment of fecal samples were performed as mentioned above. RNA viruses in samples were extracted by using Magen RNA extraction kits (Magen, China), and the whole genome was amplified with Qiagen kit (REPLI-g Cell WGA & WTA Kit).
Sequencing libraries construction and sequencing method were identical as DNA virome. The bioinformatics analyses were similar as mentioned above. Specifically, Blast software (v2.9.0 +) was used to compare the unique contig with virus database, if the alignment similarity was ≥ 80%, the alignment length was ≥ 500 bp, and e ≤ 1e-5 was defined as virus sequence; if the alignment length was ≥ 100 bp, and e ≤ 1e-5 was defined as suspected virus sequence. The candidate viruses sequences were queried against the NCBI reference genomes using BWA-MEM, NT database using MegaBLAST (e-value cutoff 1E-10), BLASTn (e-value cutoff 1E-10), and NCBI NR database using BLASTx (e-value cutoff 1E-3). Sequences with significant hits were classified as novel viruses based on the taxonomy identity of the best BLAST hit [40, 41]. Parts of the viruses’ contigs, such as Rotavirus, Gammacoronavirus and Deltacoronavirus were aligned using Blast software to compare their similarity with other references. MEGA 6.0 software was used to build the phylogenetic tree using N-J methods with 1000 bootstraps.
Verification of NGS data
Total genome of soil samples were extracted by using soil genomic DNA extraction kit (Tiangen, Beijing) and water samples were concentrated by using 0.45 μm filtration (Sartorius, German), followed by extracted of their genomic DNA. All environmental extracted DNA samples as well as metagenomic DNA library of seagull feces were detected for Salmonella and Shigella spp. by using real-time PCR kits (Bioperfectus, Jiangsu). The purpose of this experiment was to verify the accuracy of NGS data and rule out the possibility of environmental contamination.
In order to verify the NGS results of the novel viruses in RNA virome, we designed PCR primers for contig 2623, contig 857 and contig 137 (Additional file 7). PCR amplification was performed using 20 μl system, containing 10 μl of Taq premix, 8 μl of water, 0.5 μl of upstream and downstream primers (10 μmol), and 1 μl of cDNA library of extracted RNA. The products were observed by 1.5% agarose gel electrophoresis and sent for bidirectional sequencing to Shuoqing, Kunming. MEGA 6.0 software was used for sequence alignment.
Availability of data
All data generated or analyzed during this study are included in this published article. The sequence data have been deposited into the National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/ with BioProject accession number: PRJNA849401 for metagenomics; PRJNA847753 for DNA virome and PRJNA848403 for RNA virome.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- PCoA
Principal Co-ordinates Analysis
- NMDS
Non-metric multidimensional scaling
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- CARD
Comprehensive Antibiotic Resistance Database
- LEfSe
Linear discriminant analysis Effect Size
Authors’ contributions
B.Y. and Y.Z. designed the research; F.L., J.Q., R.Y., W.G., and R.L. performed the research; T.Y. and X.F. analyzed the data; F.L. and W.G. wrote the paper. The author(s) read and approved the final manuscript.
Funding
This work was supported by the Joint Project of Applied Basic Research of Kunming Medical University (202001AY070001-291).
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/ with BioProject accession number: PRJNA849401 for metagenomics; PRJNA847753 for DNA virome and PRJNA848403 for RNA virome.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Feng Liao, Jing Qian and Ruian Yang contributed equally to this work.
Contributor Information
Feng Liao, Email: lf_0305@163.com.
Jing Qian, Email: qianjing2210@163.com.
Ruian Yang, Email: yangruianyyn@163.com.
Wenpeng Gu, Email: gu_02788@163.com.
Rufang Li, Email: 871194351@qq.com.
Tingting Yang, Email: xiaoyvzi@126.com.
Xiaoqing Fu, Email: yncdcfxq@163.com.
Bing Yuan, Email: 506638016@qq.com.
Yunhui Zhang, Email: yunhuizhang03@163.com.
References
- 1.Li H, Chen Y, Machalaba CC, Tang H, Chmura AA, Fielder MD, Daszak P. Wild animal and zoonotic disease risk management and regulation in China: Examining gaps and One Health opportunities in scope, mandates, and monitoring systems. One Health. 2021;13:100301. doi: 10.1016/j.onehlt.2021.100301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoarau AOG, Mavingui P, Lebarbenchon C. Coinfections in wildlife: Focus on a neglected aspect of infectious disease epidemiology. PLoS Pathog. 2020;16(9):e1008790. doi: 10.1371/journal.ppat.1008790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritzsche McKay A, Hoye BJ. Are Migratory Animals Superspreaders of Infection? Integr Comp Biol. 2016;56(2):260–267. doi: 10.1093/icb/icw054. [DOI] [PubMed] [Google Scholar]
- 4.Morse SS, Mazet JA, Woolhouse M, Parrish CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI, Daszak P. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380(9857):1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M, Liu H, Bi Y, Sun J, Wong G, Liu D, Li L, Liu J, Chen Q, Wang H, et al. Highly Pathogenic Avian Influenza A(H5N8) Virus in Wild Migratory Birds, Qinghai Lake. China Emerg Infect Dis. 2017;23(4):637–641. doi: 10.3201/eid2304.161866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J, Wang Z, Du Y, Jia Y, Wang L, Xu S, Zhu Q. Clade 2.3.2.1 H5N1 avian influenza viruses circulate at the interface of migratory and domestic birds around Qinghai Lake in China. Vet Microbiol. 2019;235:234–242. doi: 10.1016/j.vetmic.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Hu X, Liu D, Wang M, Yang L, Zhu Q, Li L, Gao GF. Clade 2.3.2 avian influenza virus (H5N1), Qinghai Lake region, China, 2009–2010. Emerg Infect Dis. 2011;17(3):560–562. doi: 10.3201/eid1703.100948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao F, Gu W, Li D, Liang J, Fu X, Xu W, Duan R, Wang X, Jing H, Dai J. Characteristics of microbial communities and intestinal pathogenic bacteria for migrated Larus ridibundus in southwest China. Microbiologyopen. 2019;8(4):e00693. doi: 10.1002/mbo3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3(6):504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 10.Phan TG, Kapusinszky B, Wang C, Rose RK, Lipton HL, Delwart EL. The fecal viral flora of wild rodents. PLoS Pathog. 2011;7(9):e1002218. doi: 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvey E, Holmes EC. Diversity and evolution of the animal virome. Nat Rev Microbiol. 2022;20(6):321–334. doi: 10.1038/s41579-021-00665-x. [DOI] [PubMed] [Google Scholar]
- 12.Phan TG, Vo NP, Boros A, Pankovics P, Reuter G, Li OT, Wang C, Deng X, Poon LL, Delwart E. The viruses of wild pigeon droppings. PLoS ONE. 2013;8(9):e72787. doi: 10.1371/journal.pone.0072787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wille M, Shi M, Hurt AC, Klaassen M, Holmes EC. RNA virome abundance and diversity is associated with host age in a bird species. Virology. 2021;561:98–106. doi: 10.1016/j.virol.2021.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Z, Qiu Y, Ge X. The taxonomy, host range and pathogenicity of coronaviruses and other viruses in the Nidovirales order. Anim Dis. 2021;1(1):5. doi: 10.1186/s44149-021-00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yarza P, Yilmaz P, Pruesse E, Glockner FO, Ludwig W, Schleifer KH, Whitman WB, Euzeby J, Amann R, Rossello-Mora R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12(9):635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 16.Reperant LA, Osterhaus A. AIDS, Avian flu, SARS, MERS, Ebola, Zika... what next? Vaccine. 2017;35(35):4470–4. [DOI] [PMC free article] [PubMed]
- 17.Ramirez-Martinez LA, Loza-Rubio E, Mosqueda J, Gonzalez-Garay ML, Garcia-Espinosa G. Fecal virome composition of migratory wild duck species. PLoS ONE. 2018;13(11):e0206970. doi: 10.1371/journal.pone.0206970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarma D, Sanchez MI, Green AJ, Peralta-Sanchez JM, Hortas F, Sanchez-Melsio A, Borrego CM. Faecal microbiota and antibiotic resistance genes in migratory waterbirds with contrasting habitat use. Sci Total Environ. 2021;783:146872. doi: 10.1016/j.scitotenv.2021.146872. [DOI] [PubMed] [Google Scholar]
- 19.Monaco CL, Kwon DS. Next-generation Sequencing of the DNA Virome from Fecal Samples. Bio Protoc. 2017;7(5):e2159. [DOI] [PMC free article] [PubMed]
- 20.Merhi G, Koweyes J, Salloum T, Khoury CA, Haidar S, Tokajian S. SARS-CoV-2 genomic epidemiology: data and sequencing infrastructure. Future Microbiol. 2022;17:1001–1007. doi: 10.2217/fmb-2021-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Zhou ZJ, You Z, Wu DY, Liu SJ, Zhang WL, Fan KR, Luo R, Qiu Y, Ge XY. Epidemiology and evolution of novel deltacoronaviruses in birds in central China. Transbound Emerg Dis. 2022;69(2):632–644. doi: 10.1111/tbed.14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu KK, Zhou ZJ, Wang Q, Ye SB, Guo L, Qiu Y, Zhang YZ, Ge XY. Characterization of Deltacoronavirus in Black-Headed Gulls (Chroicocephalus ridibundus) in South China Indicating Frequent Interspecies Transmission of the Virus in Birds. Front Microbiol. 2022;13:895741. doi: 10.3389/fmicb.2022.895741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu W, Wang W, Li W, Li N, Wang Y, Zhang W, Lu C, Tong P, Han Y, Sun X, et al. New ribotype Clostridioides difficile from ST11 group revealed higher pathogenic ability than RT078. Emerg Microbes Infect. 2021;10(1):687–699. doi: 10.1080/22221751.2021.1900748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31(10):1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 25.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38(12):e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gustavsson EK, Zhang D, Reynolds RH, Garcia-Ruiz S, Ryten M. ggtranscript: an R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics. 2022;38(15):3844–6. [DOI] [PMC free article] [PubMed]
- 28.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 29.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–D592. doi: 10.1093/nar/gkac963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5(4):e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC. Uncovering Earth's virome. Nature. 2016;536(7617):425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, Zhang X, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018;7(1):1–6. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nayfach S, Camargo AP, Schulz F, Eloe-Fadrosh E, Roux S, Kyrpides NC. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol. 2021;39(5):578–585. doi: 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo J, Bolduc B, Zayed AA, Varsani A, Dominguez-Huerta G, Delmont TO, Pratama AA, Gazitua MC, Vik D, Sullivan MB, et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome. 2021;9(1):37. doi: 10.1186/s40168-020-00990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC, Li J, Cao JP, Eden JS, et al. Redefining the invertebrate RNA virosphere. Nature. 2016;540(7634):539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 41.Zhao G, Wu G, Lim ES, Droit L, Krishnamurthy S, Barouch DH, Virgin HW, Wang D. VirusSeeker, a computational pipeline for virus discovery and virome composition analysis. Virology. 2017;503:21–30. doi: 10.1016/j.virol.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article. The sequence data have been deposited into the National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/ with BioProject accession number: PRJNA849401 for metagenomics; PRJNA847753 for DNA virome and PRJNA848403 for RNA virome.
The datasets generated and/or analysed during the current study are available in the National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/ with BioProject accession number: PRJNA849401 for metagenomics; PRJNA847753 for DNA virome and PRJNA848403 for RNA virome.