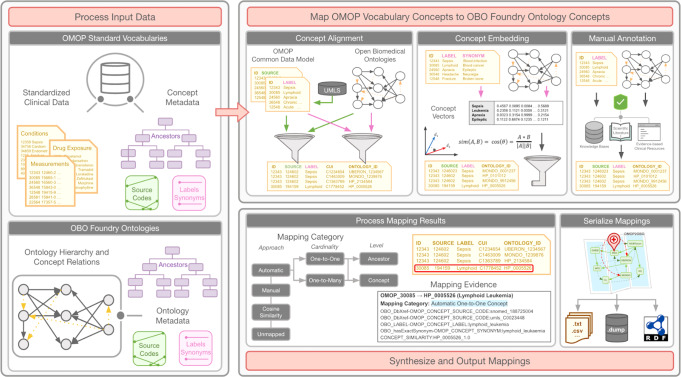
Fig. 1. Overview of the OMOP2OBO algorithm.
The OMOP2OBO algorithm consists of three components: (1) Process input data. The algorithm takes as input a table of Observational Medical Outcomes Partnership (OMOP) concepts and a list of one or more OBO (Open Biological and Biomedical Ontology) Foundry ontologies. For both data types, the algorithm expects concept or class identifiers, source codes or database cross-references, labels, synonyms, and ancestor concepts or classes. (2) Map OMOP vocabulary concepts to OBO Foundry Ontology concepts. OMOP concepts are automatically mapped to OBO Foundry ontology concepts. The algorithm includes several different approaches (e.g., concept alignment and concept embedding), prioritizing those that result in high-confidence mappings. (3) Synthesize and output mapping results. The mapping results from the prior component are post-processed to include a mapping category and human-readable evidence. Post-processed mappings are serialized and able output to a variety of file types.