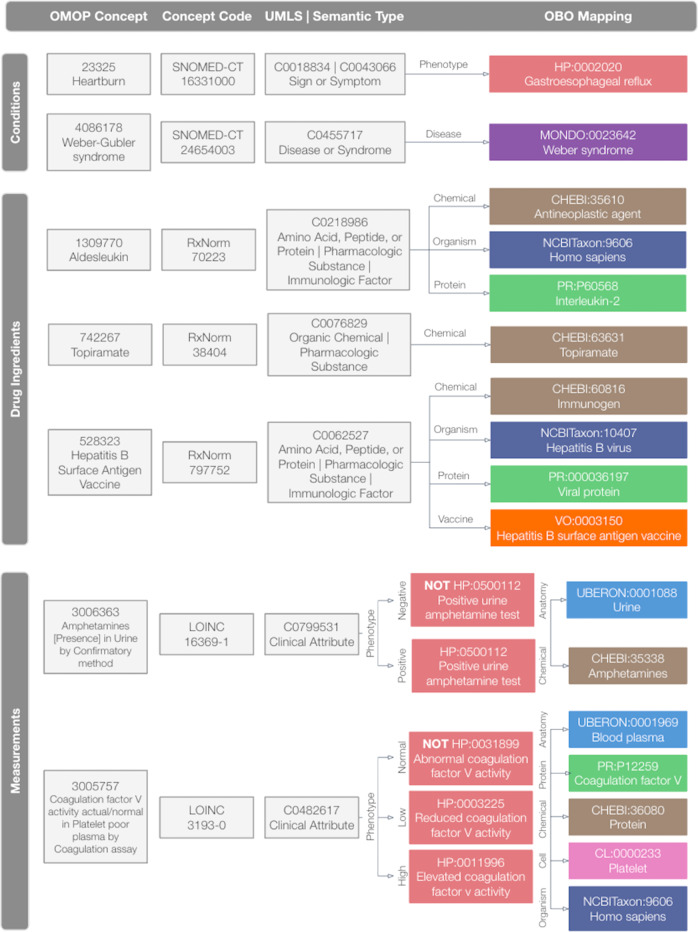
Fig. 2. OMOP2OBO mapping examples by OMOP domain.
This figure illustrates which OBO (Open Biological and Biomedical Ontology) Foundry ontologies were used for each OMOP (Observational Medical Outcomes Partnership) domain and provides example mappings. OMOP conditions were mapped to HPO and Mondo. OMOP drug ingredients were mapped to ChEBI, NCBITaxon, PRO, and VO. OMOP measurements were mapped to ChEBI, CL, HPO, NCBITaxon, PRO, and Uberon. UMLS Unified Medical Language System, HP Human Phenotype Ontology, MONDO Monarch Disease Ontology, CHEBI Chemical Entities of Biological Interest, NCBITaxon National Center for Biotechnology Information Taxon Ontology, PR Protein Ontology, VO Vaccine Ontology, UBERON Uber-Anatomy Ontology, CL Cell Ontology.