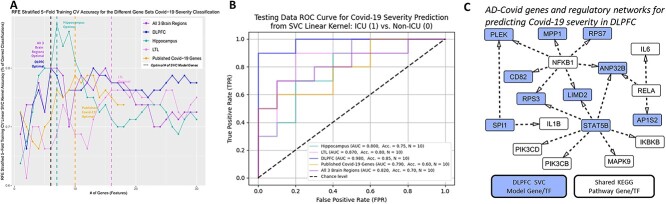
Figure 4.
Predicting Covid-19 severity using AD–Covid GRNs. (A) Prediction accuracy of Covid-19 severity after selecting different numbers of genes from AD–Covid GRNs and recently found Covid-19 genes (benchmark genes). The accuracy was calculated on the basis of the support vector machine classification (SVM or SVC) model with 5-fold stratified cross-validation on 80 balanced training samples. The dashed lines correspond to the minimal numbers of select genes with the highest accuracy (i.e. optimal gene sets for predicting Covid-19 severity). (B) Receiver operating characteristic curves and corresponding AUC values for classifying Covid-19 severity in the test data of 20 balanced samples using the SVC machine learning models. (C) Subnetwork of the DLPFC GRN relating to the 10 AD–Covid DLPFC genes for predicting Covid-19 severity (N = 10) with the shared KEGG genes. Blue: genes/TFs found in the optimal DLPFC final model (which are also 10 of the 36 AD–Covid genes). White: 1 of the 22 shared KEGG genes (between AD and Covid KEGG networks). There is no overlap between both sets of genes.