Figure 1.
Validated transcriptional programs in TC Tfh cells across platforms, gating strategies, and cohorts
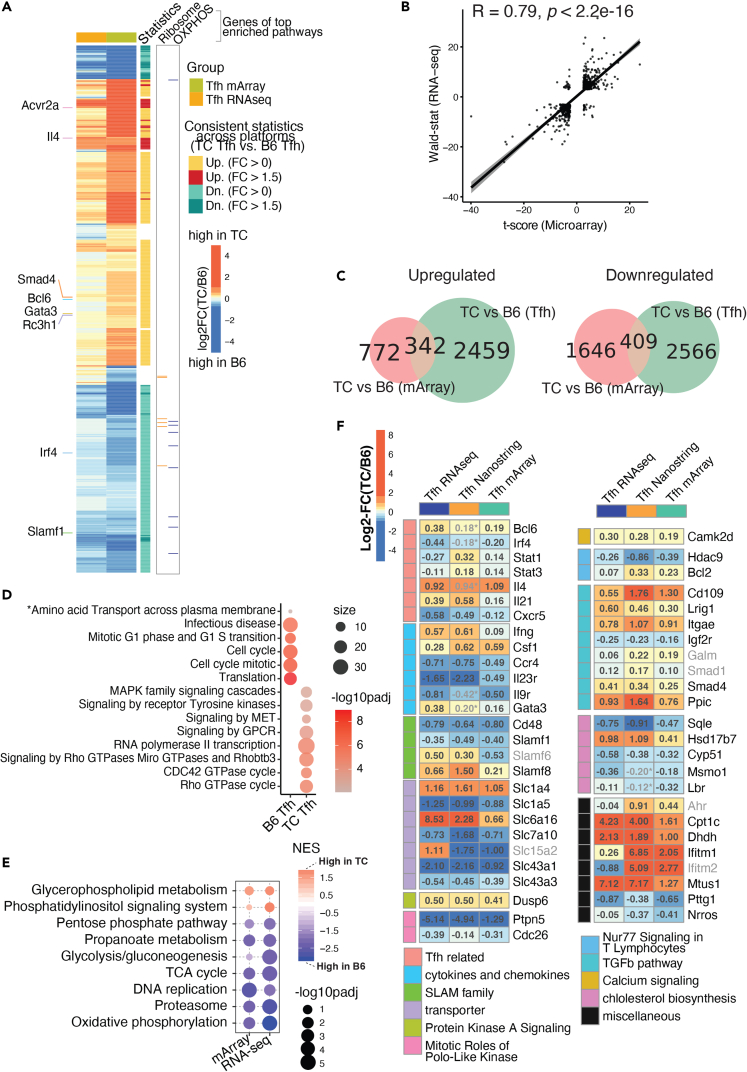
Combined analysis of the RNA-seq dataset (this report) from Tfh cells defined as CD44+ PSGL-1lo PD-1+CD4+ isolated from anti-dsDNA IgG-positive TC mice (n = 3) and B6 counterparts (n = 4) and Microarray (mArray) and Nanostring data from Tfh cells defined as PD1hi CXCR5+ CD4+ from TC (n = 5) and B6 (n = 3) mice (ref. 25).
(A) Heatmap of log2FC comparing TC vs. B6 Tfh cells computed from the mArray and RNA-seq datasets. Selected genes related to Tfh function are showed on the left. DEGs with upregulation/downregulation consistent across platforms between TC and B6 are annotated on the right along with genes of interest belonging to two metabolic pathways, OXPHOS, and ribosome, (B) Pearson correlation analysis of transcriptional profiles represented by t-scores from mArray (X axis) or Wald-statistic scores from RNA-seq (Y axis) datasets.
(C) Venn diagrams visualizing shared DEGs upregulated (left) and downregulated (right) in TC relative to B6 Tfh cells between mArray (pink) and RNA-seq (green) datasets. DEGs were filtered with FDR-corrected p < 0.05 in either direction.
(D) Gene set overlap test of the canonical pathways representing the 342 DEGs upregulated in TC and 409 DEGs upregulated in B6 Tfh cells shown in C. The color scale indicates levels of significance, and the size of the symbols indicates the number of significant genes in each pathway. The gene sets are from the canonical pathway database. The asterisk (“Amino Acid Transport”) indicates a pathway obtained from a separate analysis on DEGs filtered by FDR-corrected p < 0.05 and fold change >1.5.
(E) Significant gene sets derived from GSEA analysis comparing TC and B6 Tfh cells in either RNA-seq or mArray, filtered by FDR-corrected p < 0.25. The color scale indicates normalized enrichment scores (NES), and the size of the symbols indicates levels of significance.
(F) Heatmaps of DEGs between TC and B6 Tfh cells whose expression was concordant across three platforms, RNA-seq , mArray, and Nanostring, each in a different cohort of mice. The log2FC for each platform are shown in each cell, with the color indicating the size and direction of the change according to the scale shown in the upper left corner. DEGs were selected if they trended in the same direction between any of two assays (FDR-corrected p < 0.1) with a few exceptions with p > 0.1 labeled with asterisk (∗) and grayed out. The names of DEGs with direction consistent across only two platforms are also grayed out. DEGs were grouped by functions indicated to the left, with the corresponding keys at the bottom of the graph.