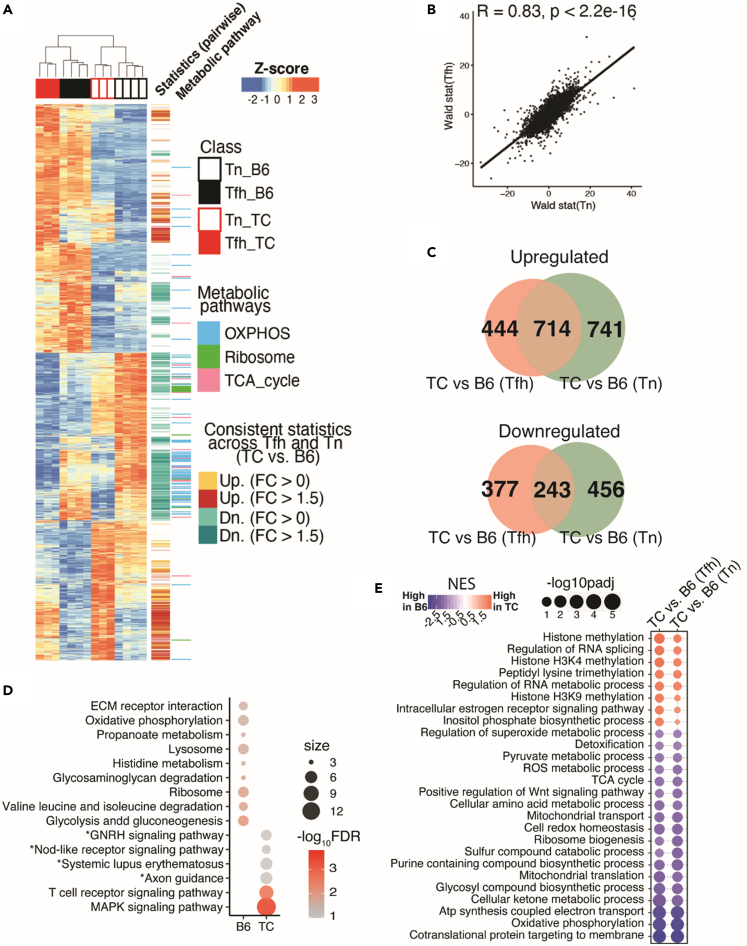
Figure 2.
Shared transcriptional signatures of TC Tn and Tfh cells relative to their B6 counterparts
(A) Heatmap showing Z-scores transformed from regularized-Log2-converted counts of the whole transcriptional profile of DEGs (FDR-corrected p < 0.05, likelihood ratio test) in the 4 groups. The track on the right shows consistently upregulated DEGs comparing TC to B6 in both Tfh and Tn cells as labeled in yellow (FC > 0) or red (FC > 1.5); the consistently downregulated counterparts are labeled in cyan (FC > 0) or dark cyan (FC > 1.5). DEGs categorized in the indicated metabolic pathways are also highlighted in the further right track.
(B) Pearson correlation analyses of transcriptional profiles comparing Wald-statistic scores of the comparison of TC Tfh vs. B6 Tfh (Y axis) and the scores of the comparisons of TC Tn vs. B6 Tn (X axis).
(C) Venn diagrams visualizing DEGs (FDR-corrected p < 0.05, FC > 1.5) comparing B6 to TC mice shared by Tfh and Tn cells. The non-shared DEGs are uniquely (bottom) or up- (top) regulated in Tfh (pink) or Tn (green) cells.
(D) Gene set overlap test of DEGs upregulated in either TC or B6 and shared by both Tfh and Tn comparisons with the same cutoff as in panel C. The color scale indicates levels of significance, and the size of the symbols indicates the number of significant genes in each pathway. Pathways are filtered by FDR <0.05, except for pathways with an asterisk (FDR <0.15). The pooled gene sets are from the KEGG database from the Molecular Signature Database (MSigDB v5.2).
(E) Bubble plot of gene sets significantly different between TC vs. B6 mice in both Tfh and Tn cells, filtered by FDR-corrected p < 0.05. The inputs are ranked gene lists based on Wald-statistic scores comparing TC vs. B6 in either Tfh or Tn cells. The gene sets are from Gene Ontology database.