Abstract

While several polyphenols were found to either inhibit or modulate the aggregation of proteins implicated in neurodegenerative diseases, such as Parkinson’s disease (PD), discrepant action mechanisms have been reported. This, in addition to some polyphenols’ pan-assay interference compounds’ reputation, casts some doubts concerning their therapeutic relevance. Here, we studied, through molecular dynamics and enhanced sampling methods, the aggregation of 11-mer peptides from the non-amyloid-β component, an aggregation-prone domain of α-synuclein (α-syn) implicated in PD and other synucleinopathies, in neat water and aqueous solutions of resveratrol (RSV) and gallic acid (GA). Further, simulations of the complete protein were carried out in aqueous urea, RSV, and GA solutions. Our results show that peptide aggregation is not disrupted by either phenolic compound. Thus, instead, intrusion of RSV and GA in the inter-peptide region induces a peptide–peptide re-orientation, favoring terminal interactions that manifest in the formation of barrierless solvent-separated configurations. Moreover, although the (poly)phenols induce a pronounced peptide dewetting at high concentrations, β-sheet-rich regions, a hallmark of α-syn aggregation, are not disrupted. Thus, our results indicate that, if anything, RSV and GA delay or modulate peptide aggregation at high concentrations via the stabilization of solvent-separated conformations as opposed to aggregation inhibition. Structural analysis of the full protein, however, shows that the (poly)phenols induce more extended conformations of α-syn, similar to urea, possibly also influencing its aggregation propensity. However, opposite to urea, the (poly)phenols reduce α-syn’s conformational space, likely due to steric effects and a slowdown of the solvent dynamics. These effects are concentration-dependent and possibly unattainable at therapeutic-relevant concentrations. These results suggest that the aggregation inhibition activity of RSV and GA in vitro should involve, instead, either the non-covalent binding to oligomeric intermediates or the stabilization of the monomer and/or oligomers through the formation of covalent bonds of the respective quinones with α-syn. In addition, the enhanced aggregation tendency of the peptides observed here could be associated with the formation of non-toxic oligomers, reported for some polyphenols.
Keywords: neurodegenerative diseases, protein aggregation, aggregation inhibitors, α-synuclein, hydrophobic effect, proteinopathies, synucleinopathies, polyphenols
1. Introduction
The etiology of neurodegenerative diseases (NDs) such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) has been linked with the formation of aberrant cytotoxic protein oligomers.1−3 A major pathological hallmark of PD, in particular, is the formation of α-synuclein (α-syn) oligomers4−7 that accumulate in intracellular inclusions called Lewy bodies and Lewy neurites,8,9 being responsible for the loss of nigral dopaminergic neurons.10,11 Notwithstanding significant advances in understanding AD, PD, and other proteinopathies, and the report of various small-molecule and peptide drugs exhibiting aggregation inhibitory activity in in vitro models, treatments hampering the formation and/or stability of these oligomers remain unavailable.12,13
Among the panoply of small molecules and peptides found to inhibit protein aggregation, polyphenols14−19 are distinguished for showing activity in several proteinopathies, including PD.20−37 The aggregation inhibitory mechanism is, however, neither always understood nor coincident among in vitro aggregation models, and some polyphenols’ neuroprotection has been linked almost exclusively to beneficial effects on concomitant pathogenic events such as oxidative stress or defective mitochondrial function.38,39
Meng et al.21 found that the ability of flavonoids to inhibit α-syn fibrillation was associated with vicinal dihydroxyphenyl moieties irrespective of the ring position where they are located. Flavonoids with three vicinal hydroxyl groups [e.g., baicalein, epigallocatechin gallate (EGCG)—see Figure 1] exhibited enhanced aggregation inhibition on α-syn fibrillation.21 Aggregation inhibition was found to occur through a combined stabilization of the monomer and soluble oligomers. Furthermore, the covalent binding of the flavonoids’ respective oxidized species (i.e., quinones) to α-syn was found to be key to the α-syn fibrillation inhibition. Caruana et al.26 also found that the main factors underpinning α-syn self-assembly inhibition and destabilization are the existence of aromatic elements that bind to the α-syn monomer/oligomer and vicinal hydroxyl groups on a single phenyl ring; compounds with three hydroxyl groups in the same phenyl ring were found to be stronger inhibitors than those with two hydroxyl groups, with the exception of nordihydroguaiaretic acid. However, the in vitro action mechanism of polyphenols seems to depend on the experimental aggregation model. Thus, it is not clear whether the inhibition/disaggregation mechanisms in the latter study26 are imparted by the respective quinones through polyphenol auto-oxidation21 and quinone-α-syn complexation or α-syn-polyphenol non-covalent interactions, instead.
Figure 1.
Structures of EGCG, curcumin, gallic acid, RSV, and baicalein.
Furthermore, whereas Caruana et al.26 found that polyphenols such as baicalein and EGCG inhibit oligomer aggregation, other studies found out that these compounds modulate aggregation, inducing the assembly of non-cytotoxic oligomers.27,28,40
Ehrnhoefer et al.27 reported that EGCG redirects amyloid fibril formation into highly stable spherical non-cytotoxic oligomers by directly binding to the natively unfolded α-syn, preventing their conversion into β-sheet-rich structures, a key step of the nucleation stage in α-syn aggregation.41 Baicalein and EGCG were also shown to disaggregate preformed α-syn amyloid fibrils.22,28
Singh et al.25 showed that curcumin (see Figure 1) does not bind to monomeric α-syn but rather to oligomeric intermediates. The proposed mechanism foresees that curcumin reduces oligomer cytotoxicity by binding to hydrophobic domains, limiting water exposure and weakening hydrophobic interactions.25 However, curcumin did not dissociate preformed amyloids into monomeric α-syn.25 A combined experimental and simulation study proposed that curcumin in combination with β-cyclodextrin (β-CD) not only inhibits aggregation but also disaggregates α-syn amyloid structures in vitro.33 Gautam et al.33 hypothesized that curcumin interacted with hydrophobic and hydrophilic middle regions of α-syn, whereas β-CD interacted with aromatic residues. Similar conclusions were extended to other polyphenols in a subsequent study,34 with varying efficiencies, namely, resveratrol (RSV), baicalein, and EGCG (see Figure 1).
In addition to NDs and proteinopathies in general, polyphenols have long been associated with positive effects on other pathologies, including the prevention of cancer and cardiovascular disease.42 However, some of these potential drugs (e.g. curcumin) have also gained the reputation of pan-assay interference compounds,43 being identified as good drug leads in assays for different pathologies.13
In light of the above, it is clear that more needs to be understood concerning polyphenols’ aggregation inhibitory activity to either pursue the development of polyphenol-based drugs or settle on their inefficiency concerning this specific mechanism. For instance, Why do some polyphenols inhibit oligomer aggregation whereas others induce/modulate oligomer aggregation in different in vitro α-syn aggregation models? Can polyphenols act like protein denaturants (e.g., urea) at high enough concentrations? Why are aromatic rings and multiple hydroxyl groups important to aggregation inhibition and disaggregation? Do polyphenols enhance the solubility of α-syn? Why do some polyphenols stabilize the α-syn monomer whereas others do not seem to interact with the monomer?
To gain insight into some of these fundamental questions, we studied here the aggregation of 11-mer peptides from the so-called non-amyloid-β component44 (NAC) peptide, a particularly amyloidogenic region of α-syn (amino acids 61–95), through molecular dynamics simulations and enhanced sampling methods.
We probed two archetypical naturally occurring phenolic compounds, namely, gallic acid (GA; a phenolic acid) and RSV (a stilbene) (see Figure 1), both well known for their antioxidant activity, linked with multiple putative health benefits.45,46 The choice of RSV was motivated by the fact that this molecule is among the smallest polyphenols, whereas GA was chosen for possessing a single aromatic ring bearing three hydroxyl groups.
RSV has been reported to inhibit the aggregation of several disease-related proteins, including transthyretin, a transport protein involved in transthyretin amyloidosis,47 P53, a tumor suppressor protein,48 islet amyloid polypeptide responsible for amyloid formation in type 2 diabetes mellitus,49 as well as to inhibit Aβ peptide fibril formation and oligomer toxicity, although not oligomerization.50−52 RSV was also found to be a mild aggregation inhibitor and a poor fibril disaggregator in a dimethyl sulfoxide-induced α-syn oligomerization in vitro model.26 This was attributed to the small number of vicinal OH groups in the phenyl rings. However, aggregation inhibition activity was found to increase with the concentration.26 GA was also reported to inhibit Aβ peptide fibril formation53 and α-syn aggregation.54
While these compounds, as most polyphenols, have low solubility, absorption, and poor bioavailability, this will not concern us here as our main goal is to understand the action mechanism of simple polyphenols in α-syn-related peptide aggregation. Thus, most of our simulations are carried out at supersaturated concentrations. Nonetheless, we stress that several chemical55 and drug delivery strategies56 have long57 been discussed to overcome the above limitations.
2. Methods
2.1. α-syn, Peptides, and (Poly)phenols
Recently, we studied58 the molecular mechanism of urea concerning the aggregation inhibition of an 11-mer peptide (85AGSIAAATGFV95) from the C-terminal domain of NAC (amino acids 61–95), referred to therein as NACterm. This segment was shown to be involved in conformations of the monomer of α-syn with the potential to inhibit aggregation.59 NACterm (see Figure 2a) comprises the largest segment (i.e., 88IAAA91) of contiguous hydrophobic amino acids and the only aromatic amino acid (Phe94) of NAC. Here, in addition to NACterm, a distinct 11-mer segment, coined60 NACore (68GAVVTGVTAVA78), was studied (see Figure 2b). NACore was chosen because of its relevance to the aggregation and cytotoxicity of α-syn60 in addition to the absence of aromatic amino acids. A slightly smaller region, encompassing residues 68–76, had been previously suggested to be pivotal to the cytotoxicity of α-syn.61 We note that various segments of the NAC have been associated with the aggregation and cytotoxicity of α-syn.59−66 In addition, N-terminal domains in the NAC flanking regions have also been associated with α-syn’s aggregation mechanism and function,67,68 including the domain comprising amino acids 46–53 where some missense mutations69−71 (e.g., A53T, E46K) associated with familial PD occur.
Figure 2.

(a) NACterm “dimer”, 85(A1G1SIA2A3A4TG2FV)95, extracted from the α-syn experimental protofibril.72 (b) NACore “dimer”, 68(G1A1V1V2T1G2V3T2A2V4A3)78, extracted from the α-syn experimental protofibril.72 Bold—hydrophobic amino acids with aliphatic side chains; Bold italic—hydrophobic amino acids with aromatic side chains. β-sheet motifs are displayed through cartoon representation—backbone hydrogen bonds are omitted for clarity.
The peptides were studied in neat water and aqueous trans-RSV and GA either with a single RSV/GA molecule or in supersaturated solutions at 298 K and 0.1 MPa; since we are not interested in the role of the carboxyl group of GA, its protonated form was used. The solubility of RSV is extremely low57 (∼0.3 mg × 100 mL–1 = 30 mg/L = 0.13 mM). The potentials of mean force (PMFs) (aka binding free energy profiles) were computed at a (RSV/W) ratio of (15:9200) molecules, which corresponds to nearly 0.09 M; PMFs at a (1:9200) ratio were also computed; however, these showed nearly no effect, suggesting that a single molecule of RSV does not significantly perturb peptide aggregation.
Addition of more than one molecule of RSV showed that RSV aggregated, as expected, under supersaturated conditions; the PMF of distinct RSV models is discussed below. The solubility of GA in water is larger73 (1.47 g × 100 mL–1 = 14.7 g/L = 86 mM). The PMFs were computed for a (GA/W) ratio of (1:9200) and (25:9200); the latter corresponds to nearly 0.15 M, thus, again at a supersaturated concentration.
MD of the full protein of α-syn was also performed in neat water and in aqueous solutions of the (poly)phenols. The RSV and GA concentrations were 42 mM (50 RSV and 50 GA molecules). However, simulations with a single RSV molecule and at 4.2 mM (5 RSV molecules), and a simulation with 100 GA molecules (84 mM), in a similar side-length box, were also performed. In addition, α-syn was simulated in an 8 M aqueous urea solution for comparison purposes.
The use of supersaturated aqueous RSV and GA solutions should maximize the potential effect of the phenols on peptide aggregation,26 assuming a monotonic increasing concentration dependence. This, in addition, promotes competition between protein-(poly)phenol and (poly)phenol-(poly)phenol interactions, mimicking, to some extent, interactions with other molecules in a cell-like environment, including crowding effects since RSV and GA can form large aggregates.
2.2. Molecular Dynamics
Molecular dynamics (MD) simulations in the isothermal-isobaric (N, p, T) ensemble of NACterm and NACore in the zwitterionic form were performed in water and aqueous (poly)phenol solutions with GROMACS.74
The T and p were controlled with the Nosé-Hoover thermostat75,76 and the Parrinello–Rahman barostat,77 and the equations of motion were solved with the Verlet leap-frog algorithm with a 2 fs time-step. Electrostatic interactions were computed via the particle-mesh Ewald (PME) method.78 A cut-off of 1 nm was used for non-bonded van der Waals and for the PME real space electrostatic interactions. Heavy atom–hydrogen covalent bonds were constrained with the LINCS algorithm.79
The full protein systems were equilibrated for 250 ns in the NpT ensemble following steepest descent energy minimization and a 500 ps equilibration period in the NVT ensemble. The trajectories were then propagated in the NpT ensemble for 1 μs in water and 500 ns in the remaining systems. A relatively long equilibration period was required to observe Rg fluctuations around an average value, with the Rg systematically decreasing throughout part of the equilibration period. Thus, the starting Rg value, corresponding to the protein in a protofibril (Rg = 3.7 nm), was found to be much larger than the average values found here for the monomer. The details about the peptides’ PMF simulations are discussed below. The secondary structure80 of the peptides and α-syn was assessed with the program DSSP.80,81
2.3. Force Field
The peptides were simulated with the AMBER99sb82 force field in TIP4P-Ew83 model water. The general AMBER force field (GAFF)84 was used to build two force fields for RSV differing in the electrostatic charges. The latter were computed using the restrained electrostatic potential (RESP)85,86 and AM1-BCC87 methods for comparison purposes. The structure of the RSV molecule was optimized at the B3LYP88/aug-cc-pvtz theoretical level, and the Merz-Kollman89 charges were computed at the HF/6-31G* theoretical level. The latter calculations were performed with the program GAUSSIAN 09.90 A CHARMM general force field (CGenFF)91 was also built for RSV using CHARMM-GUI92 for comparison purposes. The partial charges of the distinct models are given in Figure S1 and Table S1 of the Supporting Information.
RSV and GA are solids at room temperature with melting points around 260 °C. The lack of experimental data for aqueous RSV solutions, such as hydration free energies, hampers validation of the force fields, which motivated the comparison between the different force fields commonly used to model small-molecule drugs. To probe the differences between the RSV models, we calculated the PMF of a pair of RSV molecules in water (see Figure S2) through umbrella sampling (see details below); the reaction coordinate, ξ, was selected to be the center of mass distance between the RSV molecules; the MD simulations with the CGenFF were performed with the mTIP3P93 water model. In spite of the differences, the distinct models predict a contact pair minimum at similar distances (GAFF/Resp: 0.37 nm; GAFF/AM1-BCC: 0.39 nm; CGenFCC: 0.42 nm) and the absence of a solvent separated pair. Although the PMFs could not be validated against experimental data, we chose to run the remaining simulations with the GAFF/AM1-BCC force field since an intermediate aggregation propensity between the GAFF/RESP and CGenFF models is observed. Nonetheless, some calculations were also performed with CHARMM3694 and the CGenFF for comparison purposes. GA was also described by a GAFF/AM1-BCC force field.
2.4. Potentials of Mean Force
The PMFs95,96 of NACterm and NACore dimers in neat water and aqueous RSV solutions were computed through umbrella sampling.97−99 The PMF of NACterm in aqueous GA solutions was also assessed. The PMF of NACore in aqueous GA solutions was not assessed since a similar result to that found with RSV and for NACterm and GA solutions was expected based on the behavior of the latter systems. The reaction coordinate, ξ, was chosen to be the distance between the COM of the middle amino acid of NACterm (i.e., Ala) and NACore (i.e., Gly), respectively. Umbrella sampling MD were carried out for the dimers in a cubic box with PBC, large enough to allow a ξ separation ∼ 3.0 nm. Whereas the peptides’ COM was initially used, comparison between the PMFs in water and aqueous (poly)phenol solutions was found to be more difficult due to the effect of the (poly)phenols on the peptides’ structure and, therefore, on the COM. The starting conformations of the peptides were obtained from the α-syn protofibril reported by Tuttle et al.72 (PDB code: 2n0a) from solid-state NMR spectroscopy (see Figure 2).
Following steepest descent energy minimization and a 2 ns equilibration period in the NpT ensemble, the peptides were pulled away with a spring constant of 5000 kJmol–1 nm–2 and a pull rate of 0.01 nmps–1 through steered MD to generate the initial configurations. A spacing of 0.05 nm was adopted, and the umbrella sampling MD was performed for 200–250 ns after steepest descent energy minimization, a 100 ps equilibration in the NVT ensemble, and 20 ns equilibration in the NpT ensemble. The PMFs were obtained through the weighted histogram analysis method.100,101 The Bayesian bootstrap method102 was used to estimate the PMF errors. The PMFs were corrected for the entropy103 by adding the factor 2RT ln(ξ), associated with the increasing sampling volume with the ξ increase.104 The PMFs were then shifted to have a zero free energy at the longest separations.
For each system, 2–3 PMFs (i.e., 2–3 replicas) were carried out starting from different initial velocities. We found that the systems could fall either in a “high” or “low” free energy contact pair basin. Whereas for some systems, increasing sampling allowed crossing from the high energy to the low energy minima, for others, the system remained in the high energy minima up to 250 ns long umbrella sampling trajectories. However, when either the high or the low energy minima in water or aqueous (poly)phenol solutions are compared, similar conclusions are found. Thus, a nearly unchanged or slightly deeper contact pair state, favoring aggregation, and the appearance of a surprisingly long range solvent-separated ensemble of conformations, favoring disaggregation, is found for the peptides in the aqueous (poly)phenol solutions.
A similar approach was used to calculate the PMF of RSV (see Figure S2). The umbrella sampling trajectories were propagated for 25 ns after an equilibration period of 100 ps in the NVT ensemble and 2 ns in the NpT ensemble.
2.5. Solvation Free Energies
The solvation free energies, ΔGsolv, of four amino acid side chains (Ala, Val, Thr, and Phe) that comprise NACore and NACterm were computed in aqueous RSV solutions at a (RSV/W) ratio of (5:5000), which correspond to 0.05 M. These were compared with values recently reported by our group in water and 8 M aqueous urea solutions using the same force fields (i.e., AMBER99sb) and method.58 ΔGsolv were also computed at a (RSV/W) ratio of (1:10,000) with both AMBER99sb and CHARMM36.
The solvation free energies were calculated through “alchemical” free energy simulations,105 with the Bennett acceptance ratio106 method. Further details are available elsewhere.58,107,108 The side-chain analogues109 were built by replacing the Cα with an H atom with the same charge of the other H–Cβ, whereas the charge of the Cβ was changed to neutralize the side chain analogue. The remaining force field parameters were kept unchanged. ΔGSol were estimated by averaging over 2–3 alchemical simulations, starting from different initial velocities.
3. Results and Discussion
3.1. Solvation Free Energies
We start our discussion by pointing out the negligible effect of RSV on the solvation free energy of the aliphatic hydrophobic (Ala, Val) and hydrophilic (Thr) amino acid side chain analogues, opposite to urea58 (see Table 1). Thus, RSV only changes the ΔGSol of Phe/toluene, slightly favoring solvation and, therefore, solubility. The latter is expected since RSV and toluene can interact through π–π stacking, similar to RSV–RSV, inhibiting the formation of hydrogen-bonds (HBs) as proton acceptors (π...HW) with water molecules. Solvation should then be entropically (and enthalpically) favored through the release of a water molecule forming an HB with the aromatic ring; nevertheless, ΔGSol has a large uncertainty compared with the remaining side chain analogues.
Table 1. Solvation Free Energies of Ala, Val, Thr, and Phe Side Chain Analogues in Water, 8 M Aqueous Urea, and Supersaturated Aqueous RSV (GAFF/AM1-BCC) Solutions at a (RSV/W) Ratio (5:5000).
| aa/analogue | water Exp ΔGsol (kJ mol–1) | water MD ΔGsol (kJ mol–1) | RSV ΔGsol (kJ mol–1) | urea ΔGsol (kJ mol–1) |
|---|---|---|---|---|
| Ala (methane) | +8.4 | +10.6 ± 0.07 | +10.5 ± 0.1 | +11.4 ± 0.1 |
| Val (n-propane) | +8.2 | +11.1 ± 0.1 | +10.8 ± 0.2 | +10.0 ± 0.2 |
| Thr (ethanol) | –21.0 | –17.7 ± 0.1 | –17.7 ± 0.1 | –18.8 ± 0.1 |
| Phe (toluene) | –3.7 | +0.8 ± 0.1 | –0.03 ± 0.5 | –2.9(5) ± 0.2 |
The respective ΔGSol in an 8 M aqueous urea solution are also displayed in Table 1.58 Urea interacts favorably with hydrophilic and hydrophobic groups, enhancing the hydration of alkanes larger than ethane.107,110 This, in turn, strongly favors disaggregated over aggregated peptide states, suggesting that if RSV can inhibit peptide aggregation, a different mechanism should come into play.
Although not computed for an aqueous RSV solution at a (5:5000) ratio, ΔGsol for Ala/methane, Val/propane, and Thr/ethanol computed with the CHARMM36 force field at a (RSV/W) ratio (1:10,000) (see Table S2) show that CGenFF RSV also does not impact the solvation of these amino acid analogues; a similar result was found for GAFF/AM1-BCC at a (1:10,000) ratio.
3.2. Potentials of Mean Force
We now discuss the aggregation of NACterm and NACore. While aggregation inhibition of such peptides by any polyphenol does not necessarily translate into a similar behavior for α-syn or other amyloidogenic proteins, it allows the probing of whether polyphenols can inhibit aggregation by interacting with these segments of NAC.
Figure S3 displays the PMFs for NACterm in neat water and aqueous RSV and GA at a (1:9200) (poly)phenol/water ratio; no significant differences can be observed. This result motivated the use of large RSV and GA concentrations to maximize any putative effects on the free energy profiles. Figure 3a–c shows the PMFs of NACterm in neat water and supersaturated aqueous RSV and GA solutions. The PMF replicas are distinguished in “high” and “low” energy PMFs. This difference is more marked for NACterm in neat water (Figure 3a) and near the contact region, in general, with fluctuations that can be of the order of ∼2 kT to 3 kT. Block average PMFs showed that some PMFs can switch from either a high to a low energy or a low to high energy profile. For most systems, however, lower fluctuations were observed and similar block PMFs were found, suggesting convergence after ∼100 ns; running a third replica for some systems showed a profile similar to one of those obtained on the other replicas.
Figure 3.

PMFs of NACterm in (a) neat water, (b) aqueous RSV, and (c) aqueous GA solutions. The replicas are distinguished in high- and low-energy PMFs.
Nonetheless, as further discussed below, such seemingly “high” and “low” energy profiles will not influence our conclusions concerning aggregation in water and in the aqueous (poly)phenol solutions.
The most prominent feature of Figure 3 is the fact that the PMFs in aqueous (poly)phenol solutions extend through much longer distances, even precluding an accurate assessment of the zero free energy domain. The PMF, W(ξ), gives the free energy profile along a reaction coordinate, ξ96,104
| 1 |
where g(ξ) is the radial
distribution function (RDF), that is, the probability of finding the
peptides at a distance ξ, k is a constant,
and the entropic term 2RT ln(ξ) was omitted.
The PMF is, thus, defined up to a constant, k, generally
set by the RDF going to 1 when ξ → ∞, that is, G(ξ = ∞) = 0. Here, the PMF was calculated
through umbrella sampling97−99 to overcome sampling limitations
in the calculation of g(ξ). However, to estimate
the binding free energy, that is, the free energy difference between
the peptides at an infinite separation and at the equilibrium distance,
long enough distances, ξmax, must be sampled such
that 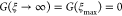 . For the aqueous RSV and GA solutions,
this distance may not have been reached and the depth of the PMFs
could be poorly estimated. To assess the PMF up to longer distances,
a larger box was used to estimate a more accurate ξmax for NACterm in an aqueous RSV solution. The discussion on the reasons
behind the peculiar long distances up to which the PMF extends is
postponed to the next sections. This simulation was carried out for
the same 15 RSV molecules and 15,500 water molecules, reducing the
concentration to 0.05 M = 50 mM, the same concentration used to compute
the solvation free energies of the amino acid side chain analogues.
. For the aqueous RSV and GA solutions,
this distance may not have been reached and the depth of the PMFs
could be poorly estimated. To assess the PMF up to longer distances,
a larger box was used to estimate a more accurate ξmax for NACterm in an aqueous RSV solution. The discussion on the reasons
behind the peculiar long distances up to which the PMF extends is
postponed to the next sections. This simulation was carried out for
the same 15 RSV molecules and 15,500 water molecules, reducing the
concentration to 0.05 M = 50 mM, the same concentration used to compute
the solvation free energies of the amino acid side chain analogues.
Figure S4 shows that whereas the PMF extends up to nearly 4.0 nm, influencing the definition of the G(ξ→∞) = 0, a similar qualitative behavior at short distances is observed, namely, a lower energy aggregate state (i.e., a contact pair) compared to that of neat water. Thus, if anything, a slightly deeper contact pair is observed when a larger value of ξmax is used.
Figure 4a–d compares the NACterm high- and low-energy PMFs, respectively, in water and aqueous RSV and GA solution. A deeper contact pair state, favoring aggregation, is found for the peptides in the aqueous RSV and GA solution. Notice that if the high- and low-energy PMFs are averaged, similar conclusions are found. We stress that this behavior was not observed for NACterm in 8 M aqueous urea solutions where a contact pair is not thermodynamically stable.58
Figure 4.
PMFs of NACterm in neat water and aqueous RSV and GA solutions.
Figure 5 shows similar plots for the NACore PMFs in water and in an aqueous RSV solution. Similar to NACterm, a deeper contact pair can be seen in the RSV solution as well as a longer tail, indicating that the PMF does not converge to zero up to ∼3 nm. When compared to NACterm, NACore is slightly more amyloidogenic in an aqueous RSV solution (see Figure S5).
Figure 5.
PMFs of NACore in neat water and aqueous RSV solutions.
To gain insight into the molecular source of the above free energy profiles, we studied the hydration, secondary structure, and the orientation of the peptides, now discussed.
3.3. Peptide Hydration
To probe the hydration level next to the peptides, hydration maps were calculated from the umbrella sampling trajectories (Figure 6). These were built by calculating the amino acids Cβ–OW (Cα–OW for glycine) and the backbone O–OW and N–OW (Figure S5) coordination numbers (CNs), along the PMF reaction path, where OW is the water molecules’ oxygen atom.
Figure 6.
Hydration maps of NACterm amino acids (Cβ) in (a) neat water, (b) an aqueous RSV solution, and (c) an aqueous GA solution from umbrella sampling simulations.
The CNs in neat water and in the RSV and GA solutions were all normalized by the maximum CNs for each amino acid in neat water
 |
2 |
where CNi is the CN of amino acid i averaged over the two peptides, g(r) is the RDF, rmin is the first minimum of the respective RDF, and CNinorm is the normalized CN for amino acid i.
Figure 6a–c shows a clear hydration/dehydration transition as the NACterm peptides approach. A similar dehydration is observed next to the backbone O and N atoms (see Figure S6). Notice that a more pronounced dehydration is expected in the central region, where interpeptide backbone contacts are maximized upon association, especially near A3, since this is used in the reaction coordinate definition. A similar result was found for NACore (see Figure S7). Figure 6 also indicates that both RSV and GA interact with the peptides, replacing water molecules, and, therefore, inducing an additional dehydration upon aggregation. Such a replacement, also observed in an 8 M aqueous urea solution,58 does not, however, inhibit aggregation, opposite to urea.
This means that (poly)phenols cannot suppress hydrophobic and electrostatic interactions, opposite to urea, a result consistent with the solvation free energies previously discussed.
We also calculated solvation maps based on the peptide–RSV and peptide–GA coordination. RSV and GA solvation maps (see Figure S8) show a lower intrusion of RSV than GA near NACterm and a lower intrusion of RSV near NACore than next to NACterm.
4. Peptides’ Relative Orientation
We now turn attention to the peculiar long-distance tail observed for the NACterm and NACore PMFs in aqueous solutions of RSV and GA. These tails indicate that the peptides still interact significantly at large values of the reaction coordinate, ξ. Such interactions must necessarily involve flanking regions of the peptides since ξ restrains any significant interaction between the central amino acid (A3) of the peptides. Since these long-range interactions are not observed in water, this indicates that a distinct relative orientation of the peptides should be induced by the (poly)phenols.
To assess the peptides’ relative orientation in neat water and in aqueous (poly)phenol solutions, the interpeptide amino acid–amino acid distances (daa) were calculated at two values of the reaction coordinate, corresponding to the contact pair (ξ = 0.55 nm) and the closest distance at which the PMF in water, but not in the (poly)phenol solutions, converges to zero (ξ = 2.05 nm).
The maps were built by calculating the average distance, daa, between the COM of each amino acid pair. The distances were then normalized by the largest daa (i.e., dmax) among every solution (e.g., neat water, RSVaq, and GAaq for NACterm) for each peptide, daa* = daa/dmax.
Figure 7 displays these maps for the NACterm in water and the aqueous solutions of RSV and GA; similar results for NACore are displayed in Figure S9. A configuration of the peptides at the respective distances is also shown in Figure 7. We note that although an antiparallel orientation was favored at ξ = 0.55 nm, observation of the trajectories at different ξ around the PMF minimum showed that the peptides can be found either in a parallel or in an antiparallel orientation.
Figure 7.
Amino acid–amino acid distance maps computed from the umbrella sampling window corresponding to the minimum (ξ = 0.5 nm) for NACterm in (a) neat water, (b) aqueous RSV solution, and (c) aqueous GA solution. The scaled reaction coordinate is ξ* = ξ/dmax = 0.26.
Although some differences are perceptible in Figure 7, especially between the maps in water and in the aqueous GA solution, a similar behavior can be seen, namely, a closer distance between the central amino acids, as expected.
On the contrary, the distance maps displayed in Figure 8 for ξ = 2.05 nm show a completely different picture in water and in the (poly)phenols’ solutions.
Figure 8.
Amino acid–amino acid distance maps computed from the umbrella sampling window corresponding to ξ = 2.05 nm for NACterm in (a) neat water, (b) an aqueous RSV solution, and (c) an aqueous GA solution. The scaled reaction coordinate is ξ* = ξ/dmax = 0.81.
Thus, while Figure 8a shows that the peptides are no longer in contact and reduced amino acid–amino acid distances vary between ∼0.8 and 1.0, Figure 8b,c shows that distances around ξ* = 0.81 are mainly observed in the central region because of the umbrella harmonic potential. Figure 8b shows that the Phe maintains a close contact with several amino acids of the opposite peptide. Figure 8c for GA shows an even more marked contact region between the peptides, with one of the peptides adopting a hairpin conformation. The hairpin tips are oriented toward one of the end regions of the opposite peptide giving rise to close contacts. Figures S9 and S10 show a similar behavior for NACore. Therefore, both RSV and GA seem to induce structural conformations that promote the peptides’ contact even at long values of the reaction coordinate, leading to a solvent-separated stabilization of the system not observed in neat water.
5. Peptides’ Secondary Structure
α-syn oligomers are characterized by the formation of interpeptide β-sheet structures. In α-syn protofibrils, these structures appear in the NAC (cross β-sheets) and N-terminal domains.72 The secondary structure of the peptides was, therefore, also assessed here. Figure 9 shows the appearance of β-sheet structures in water, upon aggregation, especially marked for NACore.
Figure 9.
Secondary structure of the peptides calculated from the umbrella sampling trajectories in water and aqueous solutions of RSV and GA; lines are a guide to the eye.
These structures, however, persist in the aqueous solutions of RSV and GA, consistent with the PMF contact pair invariance; some prominent intra-molecular β-sheets (β-hairpin) are also observed for NACore in the aqueous RSV solution. Thus, opposite to urea,58 RSV and GA do not disrupt β-sheets, indicating that any aggregation inhibition mechanism is probably not directly associated with the binding to the NAC domain of α-syn.
Although our results are limited concerning the multitude of possible binding groups in the protein, RSV and GA do not display an aggregation inhibitory activity similar to that portrayed by urea, where intrusion of enough urea molecules suppresses hydrophobic interactions and backbone HBs. Notwithstanding the above results, other aggregation inhibitory mechanisms, including a possible stabilization of α-syn, cannot be ruled out, and, therefore, such a structural mechanism was also investigated.
5.1. α-syn Structure in Aqueous Urea and (Poly)phenol Solutions
To probe the structural perturbations induced by RSV and GA, we performed MD simulations of the full protein in water and aqueous solutions of RSV and GA. These were further compared with the structure of α-syn in an 8 M aqueous urea solution. Urea is expected to induce less compact conformations of the monomer by favoring the solvation of hydrophobic amino acids.58,107,111 Furthermore, since urea inhibits aggregation, resemblances among the structure of α-syn in aqueous urea and aqueous RSV and GA solutions could indicate a similar ability to reduce its aggregation propensity. The structure of α-syn was characterized by the radius of gyration, Rg, and the secondary structure.
Figure 10a shows the distribution of Rg for α-syn in the distinct solutions. The average value of Rg in neat water is 1.6 nm. This is much lower than the experimental values (2.5–4.0 nm) inferred from different experimental techniques;41,112−115 the Rg of the starting configuration of α-syn in the protofibril is 3.7 nm. All-atom force fields such as AMBER and CHARMM are known to underpredict the Rg of IDPs in general, including α-syn, portraying a more compact, albeit, still disordered protein.116 The reason is that these force fields overestimate protein–protein and/or underestimate protein–water interactions since they were primarily developed to describe the structure of globular proteins. We note that less compact structures have been reported with some optimized coarse-grained models117,118 and modified water models.116 Nonetheless, despite the AMBER99sb limitations in reproducing the experimental Rg, we assume that the force field can qualitatively reproduce the structural transformations of α-syn in aqueous urea and (poly)phenol solutions.
Figure 10.
(a) Radius of gyration of α-syn in neat water, 42 mM aqueous RSV, 42 and 84 mM aqueous GA, and 8 M aqueous urea solutions. The Rg in the initial (experimental) configuration of the protofibril was Rg = 3.7 nm. The average Rg of AMBER99sb α-syn is 1.6 nm in water, 2.7 nm in urea, 1.8(1) nm in RSV, and 1.8(7) and 2.0(2) nm in GA; inset: radius of gyration of α-syn in RSV (1 RSV molecule, 5 RSV molecules, and 50 RSV molecules). (b) Secondary structure of α-syn in the different solutions; for GA, results for 50 GA are displayed; errors bars are negligible and were omitted for clarity. MD snapshots of α-syn in aqueous RSV, GA, and urea solutions are displayed on the right-hand side: RSV, GA, and urea molecules within 0.5 nm of the protein, respectively, are displayed; solvation water molecules are omitted for clarity.
Figure 10a shows that RSV and GA induce a less compact structure of α-syn in average, although more compact than that found in aqueous urea.
Interestingly, RSV and GA tend to narrow the Rg values accessible to the protein, nearly “freezing” the structure of α-syn, albeit only at a very high concentration. This is especially marked for aqueous GA at the highest concentration studied, with the Rg distribution approaching a delta function. This indicates that RSV and GA reduce the conformational space accessible to the protein by layering around the protein. This behavior may be associated with steric effects induced by the (poly)phenols next to the protein and/or a slowdown of the water dynamics around the protein. This is opposite to the behavior found in aqueous urea, where a broader Rg distribution is observed. Along with a smaller size, urea is known not to significantly disturb the water structure and dynamics.119,120 Our results indicate, in addition, that although the protein extends, urea still allows it to access many different conformations, opposite to RSV and GA.
The structural transformations of Figure 10a are accompanied by an increase of the random coil percentage, well correlated with the magnitude of the Rg shift (as expected) (Figure 10b). Thus, assuming that a less compact structure, as found in aqueous urea, disfavors aggregation, our results suggest that RSV and GA destabilize more aggregation-prone structures of the monomer. The magnitude of the shift is, however, relatively small at low concentrations (see Figure 10a—inset), suggesting that these molecules may not have an effective therapeutic potential via this aggregation inhibition mechanism. On the other hand, the effect of the suppression of α-syn’s conformational space on the aggregation tendency is unknown.
6. Conclusions
A panoply of polyphenols has been reported to either inhibit or modulate the aggregation of proteins associated with various proteinopathies, including α-syn, a key protein in PD, dementia with Lewy bodies, and multiple system atrophy, jointly known as synucleinopathies or Lewy body diseases. In addition, some of these polyphenols disaggregate preformed fibrils in vitro. This has granted several polyphenols, including many dietary flavonoids, the reputation of aggregation inhibitors. Furthermore, since some of these compounds showed the ability to disaggregate oligomers and mature fibrils, they could represent a pathway not only to prevent but also to reverse the disease, a pivotal issue, since NDs are often diagnosed late. The reported aggregation inhibition/modulation mechanisms, among different in vitro studies, can, however, be significantly different, and discrepant mechanisms have been put forward. The exact role of the aromatic rings and the importance of multiple hydroxyl groups in a single ring are not well-understood. Even the putative effect of these polyphenols on the solubility of the monomer, oligomers (soluble), and fibers (insoluble) remains insufficiently studied.
Herein, we studied the aggregation of two 11-mer amphiphilic segments (NACterm and NACore peptides) from NAC, a key aggregation-prone domain of α-syn, in neat water and in aqueous solutions of RSV and GA. Furthermore, the structure of the full protein was studied in aqueous solutions of the polyphenols and urea.
Our results indicate that peptide aggregation is not disrupted by either phenolic compound, even at exceedingly large concentrations, despite the polyphenols’ interaction with the protein, as opposed to remaining “trapped” in the water. Thus, instead, RSV and GA induce a peptide–peptide re-orientation, favoring terminal interactions that manifest in the formation of barrierless solvent-separated configurations extending over exceedingly large distances. Furthermore, the (poly)phenols do not disrupt β-sheet-rich regions, opposite to urea, which inhibits aggregation and disaggregates α-syn oligomers. Structural analysis of the full protein shows that although to a lower extent than urea, the (poly)phenols stabilize less compact structures of α-syn, thus possibly influencing its aggregation propensity. This stabilization is concentration-dependent and is accompanied by a reduction of the protein’s conformational space possibly due to steric effects and a slowdown of biological water, that is, water molecules sharing the protein’s surface with the (poly)phenols. These structural effects, nevertheless, seem unattainable at therapeutic relevant concentrations.
We stress that many alternative aggregation inhibition mechanisms, not assessed here, are possible. These include the aggregation inhibition via covalent binding of the respective quinones to the protein, the aggregation modulation by interacting with nascent oligomers, or the aggregation inhibition via the interaction with other amyloidogenic regions of the α-syn. Furthermore, some polyphenols have been reported to induce the formation of non-toxic oligomers. However, concerning the question on the aggregation inhibitory activity of RSV and GA of important NAC domains such as NACore and NACterm, shown to be pivotal to the aggregation process, our results indicate that these (poly)phenols do not inhibit aggregation.
Acknowledgments
G.M. acknowledges financial support from Fundação para a Ciência e a Tecnologia (FCT) of Portugal for a PhD scholarship (2021.05348.BD). C.N. acknowledges a BII (ref. 3717) scholarship from BioISI. N.G. acknowledges financial support from FCT (CEEC/2018). The work was supported by UIDB/04046/2020 and UIDP/04046/2020 centre grants from FCT, Portugal (to BioISI) and by the Portuguese National Distributed Computing Infrastructure (http://www.incd.pt). GM and N.G. acknowledge support from FCT through the computational project CPCA/A1/470255/2021.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschemneuro.3c00162.
Molecule of trans-RSV; atomic charges for different force fields of trans-RSV; potential of mean force for three force fields of RSV; solvation free energies of CHARMM36 Ala, Val, and Thr in mTIP3P water and RSV solutions; PMFs of NACterm in water and aqueous (1:9200) RSV and (1: 9200) GA solutions; PMF of NACterm in an aqueous RSV solution computed in a large box up to ∼3.75 nm; PMFs of NACterm and NACore in water and an aqueous RSV solution; hydration maps of the NACterm backbone in water and aqueous RSV solutions; hydration maps of NACore amino acids (Cβ) in water and aqueous RSV solution; NACterm and NACore, RSV and GA solvation maps; NACore aa–aa distance surfaces in water and aqueous RSV solution (ξ* = 0.21); and NACore aa–aa distance surfaces in water and aqueous RSV solution (ξ* = 0.82) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Ross C. A.; Poirier M. A. Protein Aggregation and Neurodegenerative Disease. Nat. Med. 2004, 10, S10–S17. 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Aguzzi A.; O’Connor T. Protein Aggregation Diseases: Pathogenicity and Therapeutic Perspectives. Nat. Rev. Drug Discovery 2010, 9, 237–248. 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- Sweeney P.; Park H.; Baumann M.; Dunlop J.; Frydman J.; Kopito R.; McCampbell A.; Leblanc G.; Venkateswaran A.; Nurmi A.; Hodgson R. Protein Misfolding in Neurodegenerative Diseases: Implications and Strategies. Transl. Neurodegener. 2017, 6, 6. 10.1186/s40035-017-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B.; Lansbury P. T. Separating the Responsible Protein Aggregates from The Innocent Bystanders. Annu. Rev. Neurosci. 2003, 26, 267–298. 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- Winner B.; Jappelli R.; Maji S. K.; Desplats P. A.; Boyer L.; Aigner S.; Hetzer C.; Loher T.; Vilar M.; Campioni S.; Tzitzilonis C.; Soragni A.; Jessberger S.; Mira H.; Consiglio A.; Pham E.; Masliah E.; Gage F. H.; Riek R. In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 4194–4199. 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinar D. P.; Balija M. B. G.; Kügler S.; Opazo F.; Rezaei-Ghaleh N.; Wender N.; Kim H.-Y.; Taschenberger G.; Falkenburger B. H.; Heise H.; Kumar A.; Riedel D.; Fichtner L.; Voigt A.; Braus G. H.; Giller K.; Becker S.; Herzig A.; Baldus M.; Jäckle H.; Eimer S.; Schulz J. B.; Griesinger C.; Zweckstetter M. Pre-Fibrillar α-Synuclein Variants with Impaired β-Structure Increase Neurotoxicity in Parkinson’s Disease Models. EMBO J. 2009, 28, 3256–3268. 10.1038/emboj.2009.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M. S.; Lansbury Jr P. T. Is There a Cause-and-Effect Relationship between α-Synuclein Fibrillization and Parkinson’s Disease?. Nat. Cell Biol. 2000, 2, E115–E119. 10.1038/35017124. [DOI] [PubMed] [Google Scholar]
- Spillantini M. G.; Schmidt M. L.; Lee V. M.-Y.; Trojanowski J. Q.; Jakes R.; Goedert M. α-Synuclein in Lewy bodies. Nature 1997, 388, 839–840. 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Spillantini M. G.; Crowther R. A.; Jakes R.; Hasegawa M.; Goedert M. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 6469–6473. 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E.; Rockenstein E.; Veinbergs I.; Mallory M.; Hashimoto M.; Takeda A.; Sagara Y.; Sisk A.; Mucke L. Dopaminergic Loss and Inclusion Body Formation in α-Synuclein Mice: Implications for Neurodegenerative Disorders. Science 2000, 287, 1265–1269. 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- Feany M. B.; Bender W. W. A Drosophila Model of Parkinson’s Disease. Nature 2000, 404, 394–398. 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- Stoker T. B.; Barker R. A. Recent Developments in the Treatment of Parkinson’s Disease. F1000Research 2020, 9, 862. 10.12688/f1000research.25634.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L. M.; Ashcroft A. E.; Radford S. E. Small Molecule Probes of Protein Aggregation. Curr. Opin. Chem. Biol. 2017, 39, 90–99. 10.1016/j.cbpa.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani M.; Rigacci S. Protein Folding and Aggregation into Amyloid: The Interference by Natural Phenolic Compounds. Int. J. Mol. Sci. 2013, 14, 12411–12457. 10.3390/ijms140612411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig A. J.; Derreumaux P. Inhibition of Protein Aggregation and Amyloid Formation by Small Molecules. Curr. Opin. Struct. Biol. 2015, 30, 50–56. 10.1016/j.sbi.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Singh S. K.; Dutta A.; Modi G. α-Synuclein aggregation modulation: an emerging approach for the treatment of Parkinson’s disease. Future Med. Chem. 2017, 9, 1039–1053. 10.4155/fmc-2017-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti S.; Greco C.; Tortora P.; Aprile F. Targeting Amyloid Aggregation: An Overview of Strategies and Mechanisms. Int. J. Mol. Sci. 2018, 19, 2677. 10.3390/ijms19092677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu X.-L.; Rao P. P. N.; Wang Y.-J. Anti-Amyloid Aggregation Activity of Natural Compounds: Implications for Alzheimer’s Drug Discovery. Mol. Neurobiol. 2016, 53, 3565–3575. 10.1007/s12035-015-9301-4. [DOI] [PubMed] [Google Scholar]
- Francioso A.; Punzi P.; Boffi A.; Lori C.; Martire S.; Giordano C.; D’Erme M.; Mosca L. β-Sheet Interfering Molecules Acting against β-Amyloid Aggregation and Fibrillogenesis. Bioorg. Med. Chem. 2015, 23, 1671–1683. 10.1016/j.bmc.2015.02.041. [DOI] [PubMed] [Google Scholar]
- Masuda M.; Suzuki N.; Taniguchi S.; Oikawa T.; Nonaka T.; Iwatsubo T.; Hisanaga S.; Goedert M.; Hasegawa M. Small Molecule Inhibitors of α-Synuclein Filament Assembly. Biochemistry 2006, 45, 6085–6094. 10.1021/bi0600749. [DOI] [PubMed] [Google Scholar]
- Meng X.; Munishkina L. A.; Fink A. L.; Uversky V. N. Molecular Mechanisms Underlying the Flavonoid-Induced Inhibition of α-Synuclein Fibrillation. Biochemistry 2009, 48, 8206–8224. 10.1021/bi900506b. [DOI] [PubMed] [Google Scholar]
- Meng X.; Munishkina L. A.; Fink A. L.; Uversky V. N. Effects of Various Flavonoids on the α -Synuclein Fibrillation Process. Parkinson’s Dis. 2010, 2010, 1–16. 10.4061/2010/650794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M.; Rajamani S.; Kaylor J.; Han S.; Zhou F.; Fink A. L. The Flavonoid Baicalein Inhibits Fibrillation of α-Synuclein and Disaggregates Existing Fibrils. J. Biol. Chem. 2004, 279, 26846–26857. 10.1074/jbc.M403129200. [DOI] [PubMed] [Google Scholar]
- Khuwaja G.; Khan M. M.; Ishrat T.; Ahmad A.; Raza S. S.; Ashafaq M.; Javed H.; Khan M. B.; Khan A.; Vaibhav K.; Safhi M. M.; Islam F. Neuroprotective Effects of Curcumin on 6-Hydroxydopamine-Induced Parkinsonism in Rats: Behavioral, Neurochemical and Immunohistochemical Studies. Brain Res. 2011, 1368, 254–263. 10.1016/j.brainres.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Singh P. K.; Kotia V.; Ghosh D.; Mohite G. M.; Kumar A.; Maji S. K. Curcumin Modulates α-Synuclein Aggregation and Toxicity. ACS Chem. Neurosci. 2013, 4, 393–407. 10.1021/cn3001203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana M.; Högen T.; Levin J.; Hillmer A.; Giese A.; Vassallo N. Inhibition and disaggregation of α-synuclein oligomers by natural polyphenolic compounds. FEBS Lett. 2011, 585, 1113–1120. 10.1016/j.febslet.2011.03.046. [DOI] [PubMed] [Google Scholar]
- Ehrnhoefer D. E.; Bieschke J.; Boeddrich A.; Herbst M.; Masino L.; Lurz R.; Engemann S.; Pastore A.; Wanker E. E. EGCG Redirects Amyloidogenic Polypeptides into Unstructured, off-Pathway Oligomers. Nat. Struct. Mol. Biol. 2008, 15, 558–566. 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- Bieschke J.; Russ J.; Friedrich R. P.; Ehrnhoefer D. E.; Wobst H.; Neugebauer K.; Wanker E. E. EGCG remodels mature α-synuclein and amyloid-β fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 7710–7715. 10.1073/pnas.0910723107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad B.; Lapidus L. J. Curcumin Prevents Aggregation in α-Synuclein by Increasing Reconfiguration Rate. J. Biol. Chem. 2012, 287, 9193–9199. 10.1074/jbc.M111.325548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K.; Yamada M. Antioxidant Compounds Have Potent Anti-Fibrillogenic and Fibril-Destabilizing Effects for Alpha-Synuclein Fibrils in Vitro. J. Neurochem. 2006, 97, 105–115. 10.1111/j.1471-4159.2006.03707.x. [DOI] [PubMed] [Google Scholar]
- Pandey N.; Strider J.; Nolan W. C.; Yan S. X.; Galvin J. E. Curcumin inhibits aggregation of α-synuclein. Acta Neuropathol. 2008, 115, 479–489. 10.1007/s00401-007-0332-4. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Yu Y.; Li X.; Ross C. A.; Smith W. W. Curcumin Protects against A53T Alpha-Synuclein-Induced Toxicity in a PC12 Inducible Cell Model for Parkinsonism. Pharmacol. Res. 2011, 63, 439–444. 10.1016/j.phrs.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Gautam S.; Karmakar S.; Bose A.; Chowdhury P. K. β-Cyclodextrin and Curcumin, a Potent Cocktail for Disaggregating and/or Inhibiting Amyloids: A Case Study with α-Synuclein. Biochemistry 2014, 53, 4081–4083. 10.1021/bi500642f. [DOI] [PubMed] [Google Scholar]
- Gautam S.; Karmakar S.; Batra R.; Sharma P.; Pradhan P.; Singh J.; Kundu B.; Chowdhury P. K. Polyphenols in combination with β-cyclodextrin can inhibit and disaggregate α-synuclein amyloids under cell mimicking conditions: A promising therapeutic alternative. Biochim. Biophys. Acta, Proteins Proteomics 2017, 1865, 589–603. 10.1016/j.bbapap.2017.02.014. [DOI] [PubMed] [Google Scholar]
- Borah P.; Sanjeev A.; Mattaparthi V. S. K. Computational investigation on the effect of Oleuropein aglycone on the α-synuclein aggregation. J. Biomol. Struct. Dyn. 2020, 39, 1259–1270. 10.1080/07391102.2020.1728384. [DOI] [PubMed] [Google Scholar]
- Mohammad-Beigi H.; Aliakbari F.; Sahin C.; Lomax C.; Tawfike A.; Schafer N. P.; Amiri-Nowdijeh A.; Eskandari H.; Møller I. M.; Hosseini-Mazinani M.; Christiansen G.; Ward J. L.; Morshedi D.; Otzen D. E. Oleuropein derivatives from olive fruit extracts reduce α-synuclein fibrillation and oligomer toxicity. J. Biol. Chem. 2019, 294, 4215–4232. 10.1074/jbc.RA118.005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzi L.; Bruzzone E.; Bisello G.; Leri M.; Stefani M.; Bucciantini M.; Polverino de Laureto P. Oleuropein aglycone stabilizes the monomeric α-synuclein and favours the growth of non-toxic aggregates. Sci. Rep. 2018, 8, 8337. 10.1038/s41598-018-26645-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C.-F.; Jiang L.; Zhang X.-W.; Iyaswamy A.; Li M. Resveratrol in Rodent Models of Parkinson’s Disease: A Systematic Review of Experimental Studies. Front. Pharmacol. 2021, 12, 644219. 10.3389/fphar.2021.644219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Bai L.; He J.; Zhong L.; Duan X.; Ouyang L.; Zhu Y.; Wang T.; Zhang Y.; Shi J. Recent Advances in Discovery and Development of Natural Products as Source for Anti-Parkinson’s Disease Lead Compounds. Eur. J. Med. Chem. 2017, 141, 257–272. 10.1016/j.ejmech.2017.09.068. [DOI] [PubMed] [Google Scholar]
- Hong D.-P.; Fink A. L.; Uversky V. N. Structural Characteristics of Alpha-Synuclein Oligomers Stabilized by the Flavonoid Baicalein. J. Mol. Biol. 2008, 383, 214–223. 10.1016/j.jmb.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V. N.; Li J.; Fink A. L. Evidence for a Partially Folded Intermediate in α-Synuclein Fibril Formation. J. Biol. Chem. 2001, 276, 10737–10744. 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- Vauzour D.; Rodriguez-Mateos A.; Corona G.; Oruna-Concha M. J.; Spencer J. P. E. Polyphenols and Human Health: Prevention of Disease and Mechanisms of Action. Nutrients 2010, 2, 1106–1131. 10.3390/nu2111106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell J.; Walters M. A. Chemistry: Chemical Con Artists Foil Drug Discovery. Nature 2014, 513, 481–483. 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- Uéda K.; Fukushima H.; Masliah E.; Xia Y.; Iwai A.; Yoshimoto M.; Otero D. A.; Kondo J.; Ihara Y.; Saitoh T. Molecular Cloning of CDNA Encoding an Unrecognized Component of Amyloid in Alzheimer Disease. Proc. Natl. Acad. Sci. U.S.A. 1993, 90, 11282–11286. 10.1073/pnas.90.23.11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahkeshani N.; Farzaei F.; Fotouhi M.; Alavi S. S.; Bahramsoltani R.; Naseri R.; Momtaz S.; Abbasabadi Z.; Rahimi R.; Farzaei M. H.; Bishayee A. Pharmacological Effects of Gallic Acid in Health and Disease: A Mechanistic Review. Iran. J. Basic Med. Sci. 2019, 22, 225–237. 10.22038/ijbms.2019.32806.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.-X.; Li C.-X.; Kakar M. U.; Khan M. S.; Wu P.-F.; Amir R. M.; Dai D.-F.; Naveed M.; Li Q.-Y.; Saeed M.; Shen J.-Q.; Rajput S. A.; Li J.-H. Resveratrol (RV): A Pharmacological Review and Call for Further Research. Biomed. Pharmacother. 2021, 143, 112164. 10.1016/j.biopha.2021.112164. [DOI] [PubMed] [Google Scholar]
- Sacchettini J. C.; Klabunde T.; Petrassi H. M.; Oza V. B.; Raman P.; Kelly J. W. Rational Design of Potent Human Transthyretin Amyloid Disease Inhibitors. Nat. Struct. Biol. 2000, 7, 312–321. 10.1038/74082. [DOI] [PubMed] [Google Scholar]
- Ferraz da Costa D. C.; Campos N. P. C.; Santos R. A.; Guedes-da-Silva F. H.; Martins-Dinis M. M. D. C.; Zanphorlin L.; Ramos C.; Rangel L. P.; Silva J. L. Resveratrol Prevents P53 Aggregation in Vitro and in Breast Cancer Cells. Oncotarget 2018, 9, 29112–29122. 10.18632/oncotarget.25631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R.; Sellin D.; Radovan D.; Gohlke A.; Winter R. Inhibiting Islet Amyloid Polypeptide Fibril Formation by the Red Wine Compound Resveratrol. ChemBioChem 2009, 10, 445–449. 10.1002/cbic.200800762. [DOI] [PubMed] [Google Scholar]
- Feng Y.; Wang X.; Yang S.; Wang Y.; Zhang X.; Du X.; Sun X.; Zhao M.; Huang L.; Liu R. Resveratrol Inhibits Beta-Amyloid Oligomeric Cytotoxicity but Does Not Prevent Oligomer Formation. NeuroToxicology 2009, 30, 986–995. 10.1016/j.neuro.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Ladiwala A. R. A.; Lin J. C.; Bale S. S.; Marcelino-Cruz A. M.; Bhattacharya M.; Dordick J. S.; Tessier P. M. Resveratrol Selectively Remodels Soluble Oligomers and Fibrils of Amyloid Aβ into Off-Pathway Conformers. J. Biol. Chem. 2010, 285, 24228–24237. 10.1074/jbc.M110.133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J.-F.; Qiao J.-P.; Qi C.-C.; Wang C.-W.; Zhou J.-N. The Binding of Resveratrol to Monomer and Fibril Amyloid Beta. Neurochem. Int. 2012, 61, 1192–1201. 10.1016/j.neuint.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Pukala T. L.; Musgrave I. F.; Williams D. M.; Dehle F. C.; Carver J. A. Gallic Acid Is the Major Component of Grape Seed Extract That Inhibits Amyloid Fibril Formation. Bioorg. Med. Chem. Lett. 2013, 23, 6336–6340. 10.1016/j.bmcl.2013.09.071. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Carver J. A.; Calabrese A. N.; Pukala T. L. Gallic acid interacts with α-synuclein to prevent the structural collapse necessary for its aggregation. Biochim. Biophys. Acta, Proteins Proteomics 2014, 1844, 1481–1485. 10.1016/j.bbapap.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Arbo B. D.; André-Miral C.; Nasre-Nasser R. G.; Schimith L. E.; Santos M. G.; Costa-Silva D.; Muccillo-Baisch A. L.; Hort M. A. Resveratrol Derivatives as Potential Treatments for Alzheimer’s and Parkinson’s Disease. Front. Aging Neurosci. 2020, 12, 103. 10.3389/fnagi.2020.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A.; Wang J.; Guo R.; Feng X.; Ge Y.; Liu H.; Agyei D.; Wang Q. Improving Resveratrol Bioavailability Using Water-in-Oil-in-Water (W/O/W) Emulsion: Physicochemical Stability, in Vitro Digestion Resistivity and Transport Properties. J. Funct. Foods 2021, 87, 104717. 10.1016/j.jff.2021.104717. [DOI] [Google Scholar]
- Amri A.; Chaumeil J. C.; Sfar S.; Charrueau C. Administration of Resveratrol: What Formulation Solutions to Bioavailability Limitations?. J. Controlled Release 2012, 158, 182–193. 10.1016/j.jconrel.2011.09.083. [DOI] [PubMed] [Google Scholar]
- Galamba N. Aggregation of a Parkinson’s Disease-Related Peptide: When Does Urea Weaken Hydrophobic Interactions?. ACS Chem. Neurosci. 2022, 13, 1769–1781. 10.1021/acschemneuro.2c00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoncini C. W.; Jung Y.-S.; Fernandez C. O.; Hoyer W.; Griesinger C.; Jovin T. M.; Zweckstetter M. From The Cover: Release of Long-Range Tertiary Interactions Potentiates Aggregation of Natively Unstructured -Synuclein. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 1430–1435. 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. A.; Ivanova M. I.; Sawaya M. R.; Cascio D.; Reyes F. E.; Shi D.; Sangwan S.; Guenther E. L.; Johnson L. M.; Zhang M.; Jiang L.; Arbing M. A.; Nannenga B. L.; Hattne J.; Whitelegge J.; Brewster A. S.; Messerschmidt M.; Boutet S.; Sauter N. K.; Gonen T.; Eisenberg D. S. Structure of the toxic core of α-synuclein from invisible crystals. Nature 2015, 525, 486–490. 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodles A. M.; Guthrie D. J. S.; Greer B.; Irvine G. B. Identification of the Region of Non-Aβ Component (NAC) of Alzheimer’s Disease Amyloid Responsible for Its Aggregation and Toxicity: Bioactive Region of NAC. J. Neurochem. 2001, 78, 384–395. 10.1046/j.1471-4159.2001.00408.x. [DOI] [PubMed] [Google Scholar]
- Madine J.; Doig A. J.; Middleton D. A. Design of an N-Methylated Peptide Inhibitor of α-Synuclein Aggregation Guided by Solid-State NMR. J. Am. Chem. Soc. 2008, 130, 7873–7881. 10.1021/ja075356q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson B. I.; Murray I. V. J.; Trojanowski J. Q.; Lee V. M.-Y. A Hydrophobic Stretch of 12 Amino Acid Residues in the Middle of α-Synuclein Is Essential for Filament Assembly. J. Biol. Chem. 2001, 276, 2380–2386. 10.1074/jbc.M008919200. [DOI] [PubMed] [Google Scholar]
- Du H.-N.; Tang L.; Luo X.-Y.; Li H.-T.; Hu J.; Zhou J.-W.; Hu H.-Y. A Peptide Motif Consisting of Glycine, Alanine, and Valine Is Required for the Fibrillization and Cytotoxicity of Human Alpha-Synuclein. Biochemistry 2003, 42, 8870–8878. 10.1021/bi034028+. [DOI] [PubMed] [Google Scholar]
- Eugene C.; Laghaei R.; Mousseau N. Early oligomerization stages for the non-amyloid component of α-synuclein amyloid. J. Chem. Phys. 2014, 141, 135103. 10.1063/1.4896381. [DOI] [PubMed] [Google Scholar]
- El-Agnaf O. M. A.; Paleologou K. E.; Greer B.; Abogrein A. M.; King J. E.; Salem S. A.; Fullwood N. J.; Benson F. E.; Hewitt R.; Ford K. J.; Martin F. L.; Harriott P.; Cookson M. R.; Allsop D. A strategy for designing inhibitors of α -synuclein aggregation and toxicity as a novel treatment for Parkinson’s disease and related disorders. FASEB J. 2004, 18, 1315–1317. 10.1096/fj.03-1346fje. [DOI] [PubMed] [Google Scholar]
- Doherty C. P. A.; Ulamec S. M.; Maya-Martinez R.; Good S. C.; Makepeace J.; Khan G. N.; van Oosten-Hawle P.; Radford S. E.; Brockwell D. J. A short motif in the N-terminal region of α-synuclein is critical for both aggregation and function. Nat. Struct. Mol. Biol. 2020, 27, 249–259. 10.1038/s41594-020-0384-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulamec S. M.; Maya-Martinez R.; Byrd E. J.; Dewison K. M.; Xu Y.; Willis L. F.; Sobott F.; Heath G. R.; van Oosten Hawle P.; Buchman V. L.; Radford S. E.; Brockwell D. J. Single residue modulators of amyloid formation in the N-terminal P1-region of α-synuclein. Nat. Commun. 2022, 13, 4986. 10.1038/s41467-022-32687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos M. H.; Lavedan C.; Leroy E.; Ide S. E.; Dehejia A.; Dutra A.; Pike B.; Root H.; Rubenstein J.; Boyer R.; et al. Mutation in the -Synuclein Gene Identified in Families with Parkinson’s Disease. Science 1997, 276, 2045–2047. 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Krüger R.; Kuhn W.; Müller T.; Woitalla D.; Graeber M.; Kösel S.; Przuntek H.; Epplen J. T.; Schols L.; Riess O. AlaSOPro mutation in the gene encoding α-synuclein in Parkinson’s disease. Nat. Genet. 1998, 18, 106–108. 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- Zarranz J. J.; Alegre J.; Gómez-Esteban J. C.; Lezcano E.; Ros R.; Ampuero I.; Vidal L.; Hoenicka J.; Rodriguez O.; Atarés B.; Llorens V.; Tortosa E. G.; del Ser T.; Muñoz D. G.; de Yebenes J. G. The New Mutation, E46K, of α-Synuclein Causes Parkinson and Lewy Body Dementia: New α-Synuclein Gene Mutation. Ann. Neurol. 2004, 55, 164–173. 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- Tuttle M. D.; Comellas G.; Nieuwkoop A. J.; Covell D. J.; Berthold D. A.; Kloepper K. D.; Courtney J. M.; Kim J. K.; Barclay A. M.; Kendall A.; Wan W.; Stubbs G.; Schwieters C. D.; Lee V. M. Y.; George J. M.; Rienstra C. M. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 2016, 23, 409–415. 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalkowsky S. H.Handbook of Aqueous Solubility Data, 2nd ed.; CRC Press: Boca Raton, FL, 2010. [Google Scholar]
- Van Der Spoel D.; Lindahl E.; Hess B.; Groenhof G.; Mark A. E.; Berendsen H. J. C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Nosé S. A Unified Formulation of the Constant Temperature Molecular Dynamics Methods. J. Chem. Phys. 1984, 81, 511–519. 10.1063/1.447334. [DOI] [Google Scholar]
- Hoover W. G. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A 1985, 31, 1695–1697. 10.1103/PhysRevA.31.1695. [DOI] [PubMed] [Google Scholar]
- Parrinello M.; Rahman A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. 10.1063/1.328693. [DOI] [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M. L.; Darden T.; Lee H.; Pedersen L. G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. 10.1063/1.470117. [DOI] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J. C.; Fraaije J. G. E. M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. . [DOI] [Google Scholar]
- Kabsch W.; Sander C. Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers 1983, 22, 2577–2637. 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Touw W. G.; Baakman C.; Black J.; te Beek T. A.; Krieger E.; Joosten R. P.; Vriend G. A Series of PDB-Related Databanks for Everyday Needs. Nucleic Acids Res. 2015, 43, D364–D368. 10.1093/nar/gku1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V.; Abel R.; Okur A.; Strockbine B.; Roitberg A.; Simmerling C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins: Struct., Funct., Bioinf. 2006, 65, 712–725. 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn H. W.; Swope W. C.; Pitera J. W.; Madura J. D.; Dick T. J.; Hura G. L.; Head-Gordon T. Development of an Improved Four-Site Water Model for Biomolecular Simulations: TIP4P-Ew. J. Chem. Phys. 2004, 120, 9665–9678. 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- Wang J.; Wolf R. M.; Caldwell J. W.; Kollman P. A.; Case D. A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Bayly C. I.; Cieplak P.; Cornell W.; Kollman P. A. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges: The RESP Model. J. Phys. Chem. 1993, 97, 10269–10280. 10.1021/j100142a004. [DOI] [Google Scholar]
- Cornell W. D.; Cieplak P.; Bayly C. I.; Kollman P. A. Application of RESP Charges to Calculate Conformational Energies, Hydrogen Bond Energies, and Free Energies of Solvation. J. Am. Chem. Soc. 1993, 115, 9620–9631. 10.1021/ja00074a030. [DOI] [Google Scholar]
- Jakalian A.; Jack D. B.; Bayly C. I. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: II. Parameterization and Validation. J. Comput. Chem. 2002, 23, 1623–1641. 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- Becke A. D. Density-functional Thermochemistry. III. The Role of Exact Exchange. J. Chem. Phys. 1993, 98, 5648–5652. 10.1063/1.464913. [DOI] [Google Scholar]
- Singh U. C.; Kollman P. A. An Approach to Computing Electrostatic Charges for Molecules. J. Comput. Chem. 1984, 5, 129–145. 10.1002/jcc.540050204. [DOI] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Petersson G. A.; Nakatsuji H.; Li X.; Caricato M.; Marenich A.; Bloino J.; Janesko B. G.; Gomperts R.; Mennucci B.; Hratchian H. P.; Ortiz J. V.; Izmaylov A. F.; Sonnenberg J. L.; Williams-Young D.; Ding F.; Lipparini F.; Egidi F.; Goings J.; Peng B.; Petrone A.; Henderson T.; Ranasinghe D.; Zakrzewski V. G.; Gao J.; Rega N.; Zheng G.; Liang W.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Throssell K.; Montgomery J. A. Jr.; Peralta J. E.; Ogliaro F.; Bearpark M.; Heyd J. J.; Brothers E.; Kudin K. N.; Staroverov V. N.; Keith T.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Millam J. M.; Klene M.; Adamo C.; Cammi R.; Ochterski J. W.; Martin R. L.; Morokuma K.; Farkas O.; Foresman J. B.; Fox D. J.. Gaussian 09, Revision D.01; Gaussian, Inc.: Wallingford CT, 2016. [Google Scholar]
- Vanommeslaeghe K.; Raman E. P.; MacKerell A. D. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of Bonded Parameters and Partial Atomic Charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S.; Kim T.; Iyer V. G.; Im W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- MacKerell A. D.; Bashford D.; Bellott M.; Dunbrack R. L.; Evanseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; Joseph-McCarthy D.; Kuchnir L.; Kuczera K.; Lau F. T. K.; Mattos C.; Michnick S.; Ngo T.; Nguyen D. T.; Prodhom B.; Reiher W. E.; Roux B.; Schlenkrich M.; Smith J. C.; Stote R.; Straub J.; Watanabe M.; Wiórkiewicz-Kuczera J.; Yin D.; Karplus M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Best R. B.; Zhu X.; Shim J.; Lopes P. E. M.; Mittal J.; Feig M.; MacKerell A. D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ 1 and χ 2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood J. G. Statistical Mechanics of Fluid Mixtures. J. Chem. Phys. 1935, 3, 300–313. 10.1063/1.1749657. [DOI] [Google Scholar]
- Chandler D.Introduction to Modern Statistical Mechanics; Oxford University Press: New York, 1987. [Google Scholar]
- Torrie G. M.; Valleau J. P. Nonphysical Sampling Distributions in Monte Carlo Free-Energy Estimation: Umbrella Sampling. J. Comput. Phys. 1977, 23, 187–199. 10.1016/0021-9991(77)90121-8. [DOI] [Google Scholar]
- Torrie G. M.; Valleau J. P. Monte Carlo Free Energy Estimates Using Non-Boltzmann Sampling: Application to the Sub-Critical Lennard-Jones Fluid. Chem. Phys. Lett. 1974, 28, 578–581. 10.1016/0009-2614(74)80109-0. [DOI] [Google Scholar]
- Kästner J. Umbrella Sampling. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2011, 1, 932–942. 10.1002/wcms.66. [DOI] [Google Scholar]
- Kumar S.; Rosenberg J. M.; Bouzida D.; Swendsen R. H.; Kollman P. A. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules. I. The Method. J. Comput. Chem. 1992, 13, 1011–1021. 10.1002/jcc.540130812. [DOI] [Google Scholar]
- Souaille M.; Roux B. Extension to the Weighted Histogram Analysis Method: Combining Umbrella Sampling with Free Energy Calculations. Comput. Phys. Commun. 2001, 135, 40–57. 10.1016/S0010-4655(00)00215-0. [DOI] [Google Scholar]
- Hub J. S.; de Groot B. L.; van der Spoel D. G_wham—A Free Weighted Histogram Analysis Implementation Including Robust Error and Autocorrelation Estimates. J. Chem. Theory Comput. 2010, 6, 3713–3720. 10.1021/ct100494z. [DOI] [Google Scholar]
- Neumann R. M. Entropic Approach to Brownian Movement. Am. J. Phys. 1980, 48, 354–357. 10.1119/1.12095. [DOI] [Google Scholar]
- Trzesniak D.; Kunz A.-P. E.; van Gunsteren W. F. A Comparison of Methods to Compute the Potential of Mean Force. ChemPhysChem 2007, 8, 162–169. 10.1002/cphc.200600527. [DOI] [PubMed] [Google Scholar]
- Duarte Ramos Matos G.; Kyu D. Y.; Loeffler H. H.; Chodera J. D.; Shirts M. R.; Mobley D. L. Approaches for Calculating Solvation Free Energies and Enthalpies Demonstrated with an Update of the FreeSolv Database. J. Chem. Eng. Data 2017, 62, 1559–1569. 10.1021/acs.jced.7b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C. H. Efficient Estimation of Free Energy Differences from Monte Carlo Data. J. Comput. Phys. 1976, 22, 245–268. 10.1016/0021-9991(76)90078-4. [DOI] [Google Scholar]
- Xavier P.; Galamba N. Effect of Urea on the Hydration and Aggregation of Hydrophobic and Amphiphilic Solute Models: Implications to Protein Aggregation. J. Chem. Phys. 2021, 155, 144501. 10.1063/5.0064707. [DOI] [PubMed] [Google Scholar]
- Galamba N. Free Energy Convergence in Short- and Long-Length Hydrophobic Hydration. J. Mol. Liq. 2021, 339, 116699. 10.1016/j.molliq.2021.116699. [DOI] [Google Scholar]
- Shirts M. R.; Pitera J. W.; Swope W. C.; Pande V. S. Extremely Precise Free Energy Calculations of Amino Acid Side Chain Analogs: Comparison of Common Molecular Mechanics Force Fields for Proteins. J. Chem. Phys. 2003, 119, 5740–5761. 10.1063/1.1587119. [DOI] [Google Scholar]
- Watlaufer D. B.; Malik S. K.; Stoller L.; Coffin R. L. Nonpolar Group Participation in the Denaturation of Proteins by Urea and Guanidinium Salts. Model Compound Studies. J. Am. Chem. Soc. 1964, 86, 508–514. 10.1021/ja01057a045. [DOI] [Google Scholar]
- Hua L.; Zhou R.; Thirumalai D.; Berne B. J. Urea Denaturation by Stronger Dispersion Interactions with Proteins than Water Implies a 2-Stage Unfolding. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 16928–16933. 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-Y.; Heise H.; Fernandez C. O.; Baldus M.; Zweckstetter M. Correlation of Amyloid Fibril β-Structure with the Unfolded State of α-Synuclein. ChemBioChem 2007, 8, 1671–1674. 10.1002/cbic.200700366. [DOI] [PubMed] [Google Scholar]
- Schwalbe M.; Ozenne V.; Bibow S.; Jaremko M.; Jaremko L.; Gajda M.; Jensen M. R.; Biernat J.; Becker S.; Mandelkow E.; Zweckstetter M.; Blackledge M. Predictive Atomic Resolution Descriptions of Intrinsically Disordered hTau40 and α-Synuclein in Solution from NMR and Small Angle Scattering. Structure 2014, 22, 238–249. 10.1016/j.str.2013.10.020. [DOI] [PubMed] [Google Scholar]
- Morar A. S.; Olteanu A.; Young G. B.; Pielak G. J. Solvent-induced collapse of α-synuclein and acid-denatured cytochrome c. Protein Sci. 2008, 10, 2195–2199. 10.1110/ps.24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Späth S.; Soranno A.; Hirschfeld V.; Hofmann H.; Rüegger S.; Reymond L.; Nettels D.; Schuler B. Charge Interactions Can Dominate the Dimensions of Intrinsically Disordered Proteins. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 14609–14614. 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piana S.; Donchev A. G.; Robustelli P.; Shaw D. E. Water Dispersion Interactions Strongly Influence Simulated Structural Properties of Disordered Protein States. J. Phys. Chem. B 2015, 119, 5113–5123. 10.1021/jp508971m. [DOI] [PubMed] [Google Scholar]
- Yu H.; Han W.; Ma W.; Schulten K. Transient β -Hairpin Formation in α -Synuclein Monomer Revealed by Coarse-Grained Molecular Dynamics Simulation. J. Chem. Phys. 2015, 143, 243142. 10.1063/1.4936910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramis R.; Ortega-Castro J.; Casasnovas R.; Mariño L.; Vilanova B.; Adrover M.; Frau J. A Coarse-Grained Molecular Dynamics Approach to the Study of the Intrinsically Disordered Protein α-Synuclein. J. Chem. Inf. Model. 2019, 59, 1458–1471. 10.1021/acs.jcim.8b00921. [DOI] [PubMed] [Google Scholar]
- Rezus Y. L. A.; Bakker H. J. Effect of Urea on the Structural Dynamics of Water. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 18417–18420. 10.1073/pnas.0606538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu A.; Fumino K.; Yukiyasu K.; Taniguchi Y. NMR Studies on Dynamic Behavior of Water Molecule in Aqueous Denaturant Solutions at 25 °C: Effects of Guanidine Hydrochloride, Urea and Alkylated Ureas. J. Mol. Liq. 2000, 85, 269–278. 10.1016/S0167-7322(00)00106-9. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.










