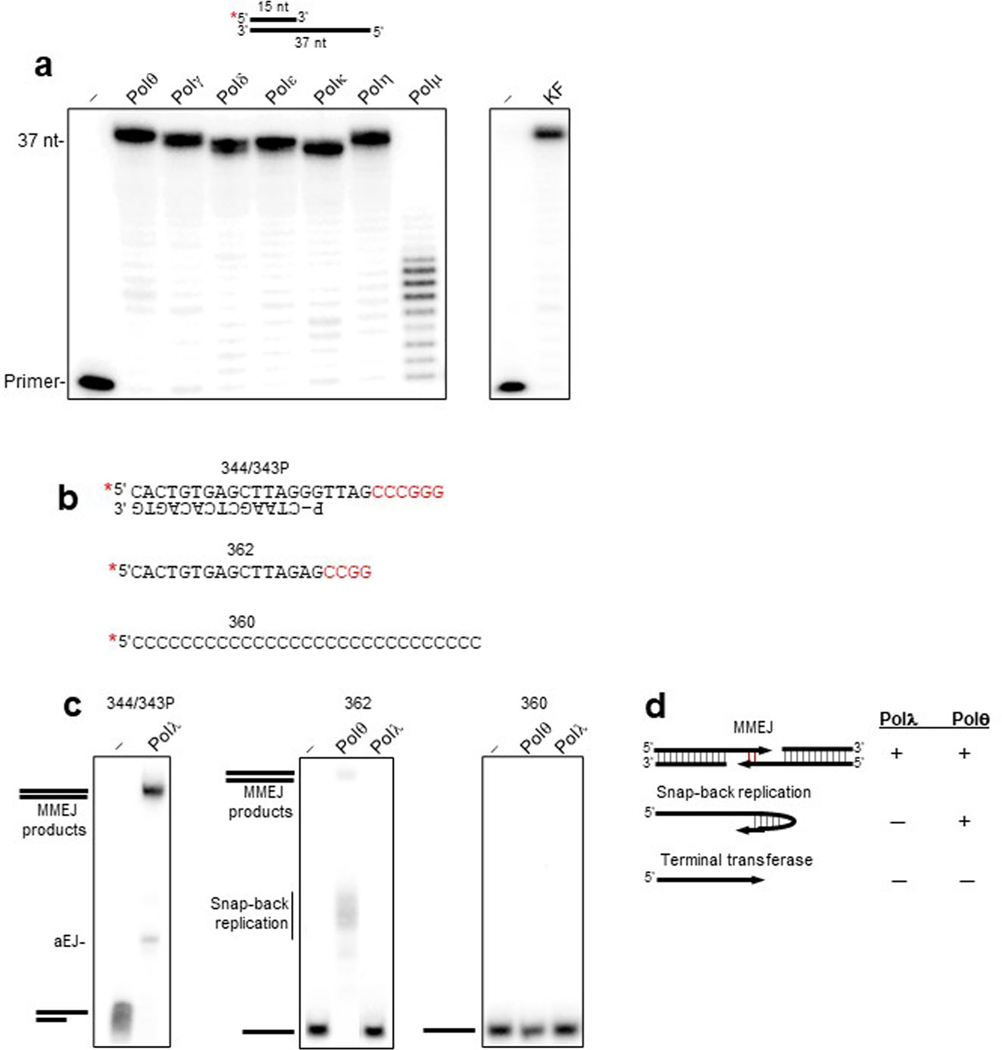
Extended Data Fig. 1 |. Controls for Pol activity on primer-templates and substrates containing 3’ ssDNA.
a, Denaturing gels showing extension of the indicated primer-template by the indicated Pols using identical conditions. Polμ is known to require a downstream ssDNA strand for optimal activity primer extension activity along a short gap. 20 nM Pol concentrations were used. b, Schematic of DNA templates. Microhomology indicated as red text. c, Non-denaturing gel showing Polλ MMEJ as a positive control for its activity (left panel). Denaturing gels showing extension of the indicated templates in the presence of all 4 dNTPs by the indicated Pols (middle and right panel). 20 nM Pol concentrations were used. d, Schematic showing the respective activities of Polθ and Polλ on the indicated templates. although both enzymes perform MMEJ (top), only Polθ exhibits ssDNA extension due to its snap-back replication activity.