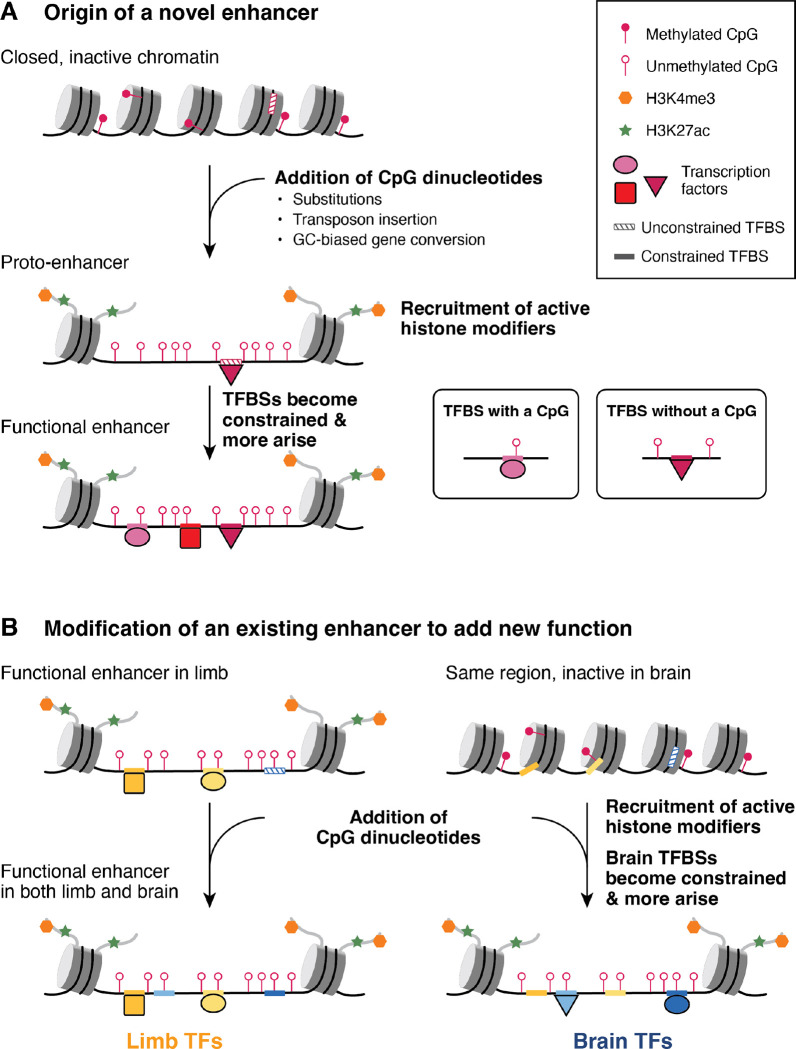
Figure 8. Model of enhancer evolution via oCGI turnover.
(A) Evolution of a new enhancer from a locus in a closed chromatin state. This locus may include unconstrained, inaccessible TFBSs (striped boxes on DNA). DNA is depicted as a black line wrapped around cylindrical nucleosomes. After oCGI gain by several potential mechanisms (indicated in the figure), the site now acts as a proto-enhancer located within open, active chromatin recruited by the oCGI (44), which allows TFs to bind previously inaccessible TFBSs. A subset of histone tails (curved gray lines) with H3K4me3 (orange hexagons) and H3K27ac (green stars) modifications are shown. Filled lollipops indicate methylated CpGs, and unfilled lollipops indicate unmethylated CpGs. Over time, TFBSs become constrained (filled boxes on DNA) and additional TFBSs may arise and become fixed, resulting in the evolution of an enhancer with a constrained biological function. (B) Co-option of an existing enhancer in a novel biological context via oCGI gain. In an ancestral species, the enhancer is active in the developing limb and inactive in the developing brain, where the chromatin at the locus is closed. After oCGI gain, CpG-related mechanisms generate open chromatin in the developing brain, which allows existing unconstrained brain TFBSs to be bound. Over time, these and additional TFBSs may gain biological functions and be maintained by selection. The locus becomes a functional enhancer in the developing brain.