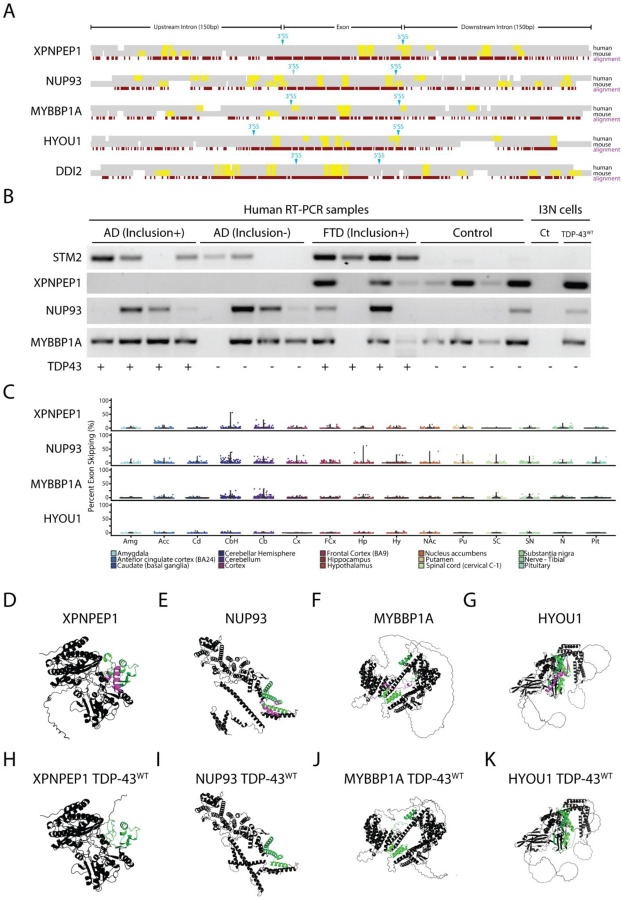
Figure 3. TDP-43 exon skipping events are found in aging human brains but do not correlate with disease.
(A) Alignment of syntenic mouse (mm10) and human (hg38) genomic sequences surrounding exons repressed by TDP-43 overexpression in human cells. Constitutively spliced exons in the genes XPNPEP1, NUP93, MYBBP1A, and HYOU1 are skipped in human cells but not mouse cells when TDP-43 is overexpressed. By contrast, the exon in DDI2 is repressed in both mouse and human cells. An analysis of UG motifs (yellow highlights) reveals that slightly higher frequencies in UG repeat frequency surrounding the 3’ and 5’ splice sites may account for species-specific exon skipping. (B) We performed single-band RT-PCR to amplify cryptic junctions or exon-repressed junctions in human brain samples from patients with AD pathology with or without TDP-43 inclusions, frontotemporal dementia with inclusions, and control patients who did not have TDP-43 inclusions. As previously reported, the cryptic exon in the gene STMN2 is highly correlated with TDP-43 proteinopathy (16, 17). RT-PCR analysis showed that exon skipping occurred in both control and disease samples, indicating that aberrant exon skipping does not correlate with TDP-43 proteinopathy. (C) Since aging is a primary factor for developing neurodegenerative disease, we wanted to explore whether skipping events appear normally in the CNS and whether skipping events correlated with aging. We analyzed publicly available human RNA-Seq datasets (GTEx) and measured the percent spliced-in values of skipping events in patients aged between 60 to 69 years old. Exon skipping was found at low levels in most of the different brain areas analyzed, with slightly higher levels in the cerebellum. Using AlphaFold 2, we modeled protein structures with (D-G) and without (H-K) exons repressed by TDP-43 overexpression. Purple highlights indicate the repressed exons while green highlights indicate flanking amino acid sequences. TDP-43 induced exon skipping can dramatically influence protein structure and may lead to functional deficits.