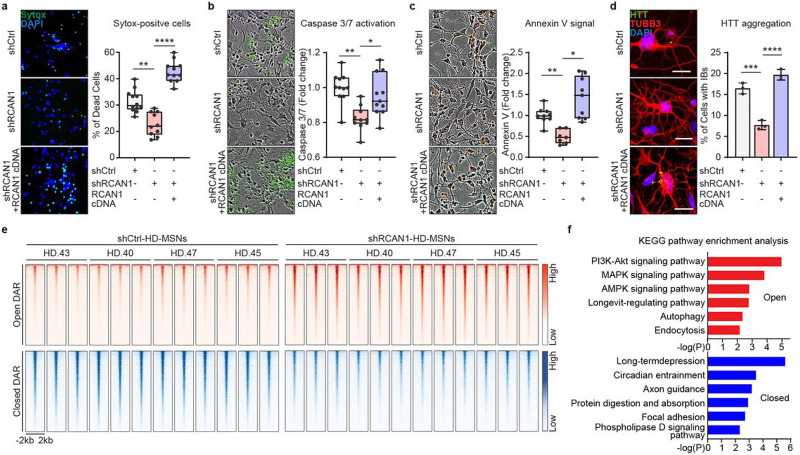
Fig. 2 ∣. RCAN1 KD protects HD-MSNs from degeneration and induces chromatin accessibility changes.
a-c, Representative images (left) and quantification (right) of Sytox-positive cells (a), Caspase 3/7 activation (green) (b), and Annexin V signal (red) (c) in three independent HD-MSNs (HD.40, HD.43, HD.47, n=10~12, independent reprogramming experiments) transduced with shControl (shCtrl), shRCAN1, or RCAN1 cDNA. d, Representative images (left) and quantification (right) of cells with HTT inclusion bodies (IBs) in three independent HD-MSNs (HD.40, HD.43, HD.47, n=3) transduced with shCtrl, shRCAN1, or RCAN1. Cells were immunostained with anti-HTT and TUBB3 antibodies. An average of 120 cells per each were counted from three or more randomly chosen fields. Scale bars represent 20 μm. e and f, Analysis of ATAC-sequencing in four independent HD-MSNs (HD.43, HD.40, HD.47, HD.45) transduced with shCtrl or shRCAN1. Heatmaps of signal intensity (e) in chromatin peaks (FDR<0.05, ∣FC∣≥1.5) of open and closed DARs in shRCAN1-HD-MSNs compared to shCtrl-HD-MSNs. KEGG pathway enrichment analysis (f) of genes associated with open (top) and closed (bottom) DARs in shRCAN1-HD-MSNs. Statistical significance was determined using one-way ANOVA (a-d); ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05, and mean±s.e.m. The sample size (n) corresponds to the number of biological replicates (a-d).