Table 2.
Kinetic parameters of various nucleobase substrates.
| Compound | Km, μM | kcata (×10−3) s−1 | kcat/Kma (×10−3) s−1 μM−1 | Ref. |
|---|---|---|---|---|
| Guanine | 2.2 | 61a | 28 a | Hoops et al. (1995) |
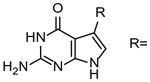
|
||||
| −CH2NH2 (preQ1) | 0.39 | 45a | 115a | Hoops et al. (1995) |
| −CN (preQ0) | 2.35 | 72a | 31 a | Hoops et al. (1995) |
| −CH2OH | 23.0 | 45a | 2.0a | Hoops et al. (1995) |
| −H | 172 | 45a | 0.3a | Hoops et al. (1995) |
| −CH3 | 255 | 45a | 0.2a | Hoops et al. (1995) |
| −CH2OCH3 | 57 | 47a | 0.8a | Hoops et al. (1995) |
| −CH2N(CH3)2 | 75 | 43a | 0.6a | Hoops et al. (1995) |
| −CONH2 | 26 | 69a | 2.7a | Hoops et al. (1995) |
| −CO2CH3 | 87 | 73a | 0.8a | Hoops et al. (1995) |
| −COCH3 | 26 | 73a | 2.8a | Hoops et al. (1995) |
| −CHO | 22 | 73a | 3.3a | Hoops et al. (1995) |
| −CO2H | 126 | 21a | 0.2a | Hoops et al. (1995) |
| −(PEG)3-thiazole orange | 9.8 | 1.6 | 0.2 | Alexander et al. (2015) |
| −C6H12-thiazole orange derivative | 1.6 | 26.7 | 16.7 | Zhou et al. (2017) |
kcat were calculated from reported Vmax values using the reported enzyme concentration (50 nM), and reaction volume (400 μL) (Hoops et al., 1995).
