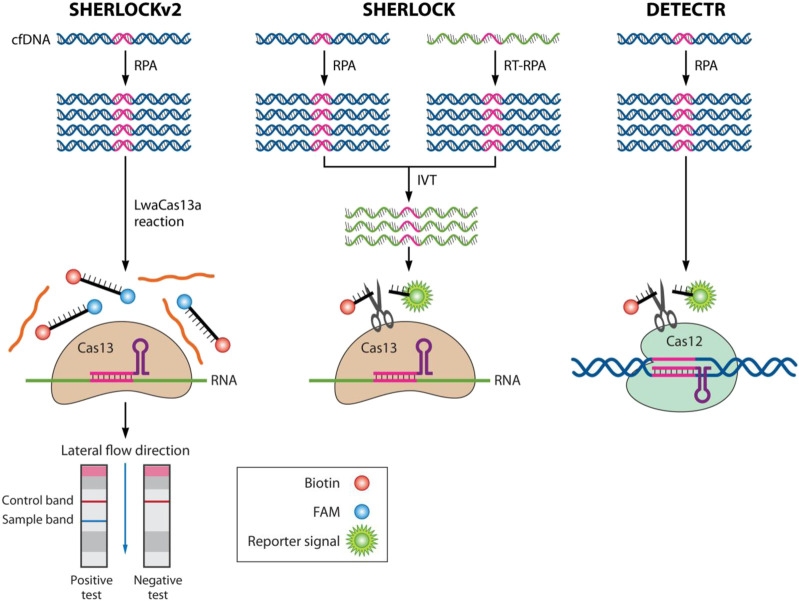
FIGURE 2.
CRISPR-Cas technologies for nucleic acid detection in SHERLOCKv2, SHERLOCK, and DETECTR assays. In the absence of its nucleic acid target, the Cas nuclease is inactive. When binding to its guide crRNA to a related target (RNA for Cas13a, ssDNA or dsDNA for Cas12a), the nuclease is activated, leading to catalytic cleavage of off-target nucleic acids (RNA for Cas13a, ssDNA for Cas12a). This collateral nuclease activity is turned into an amplified signal by providing reporter probes with a fluorophore (green) linked to a quencher (white) by a short oligonucleotide (black). (Left) Schematic of SHERLOCKv2, with direct detection of viral infection (for example) in bodily fluids. (Middle) Schematic of the SHERLOCK system. Nucleic acid is extracted from clinical samples (for example), and the target is amplified by recombinase polymerase amplification (RPA) with either RNA or DNA as the input (reverse transcriptase recombinase polymerase amplification [RT-RPA] or RPA, respectively). RPA products are detected in a reaction mixture containing T7 RNA polymerase, Cas13, a target-specific crRNA, and an RNA reporter that fluoresces when cleaved. (Right) Schematic of the DETECTR system. Reprinted with permission from Mustafa and Makhawi (2021). Copyright © 2021 Mustafa and Makhawi.