Figure 5.
Molecular modeling of compound-selected Plasmodium proteasome mutations
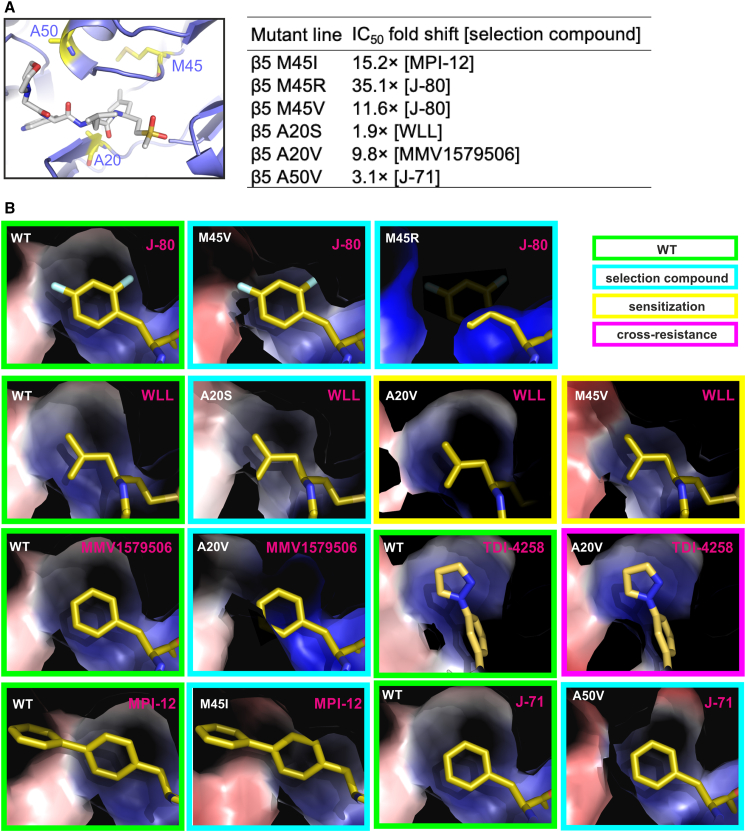
(A) Locations of the β5 A20, M45, and A50 residues (shown as sticks with yellow backbone) in the structure of the wild-type Plasmodium proteasome (shown as cartoon), with fitted WLL (cartoon with gray backbone).21,29 The table inset shows IC50 fold shifts for inhibitors tested against proteasome mutant lines compared with the wild-type parental line. Data for the β5 M45I line were taken from Xie et al.25
(B) Effects of the M45I, M45R, M45V, A20S, A20SV, and A50V mutations on the Plasmodium β5 P1 binding pocket. To facilitate comparison, the β5 P1 binding pocket of the wild-type proteasome (boxed in green) is shown in the same orientation as each of the selection mutants (cyan boxes). For all mutations, the proteasome models indicate that resistance to the selection compounds is primarily mediated by steric constraints that limit their access to the P1 binding site. Examples of sensitization (yellow boxes) and cross resistance (magenta boxes) are also shown. Protein models are represented as Van der Waals surfaces colored by electrostatic potential, overlaid with inhibitors.