Figure 6.
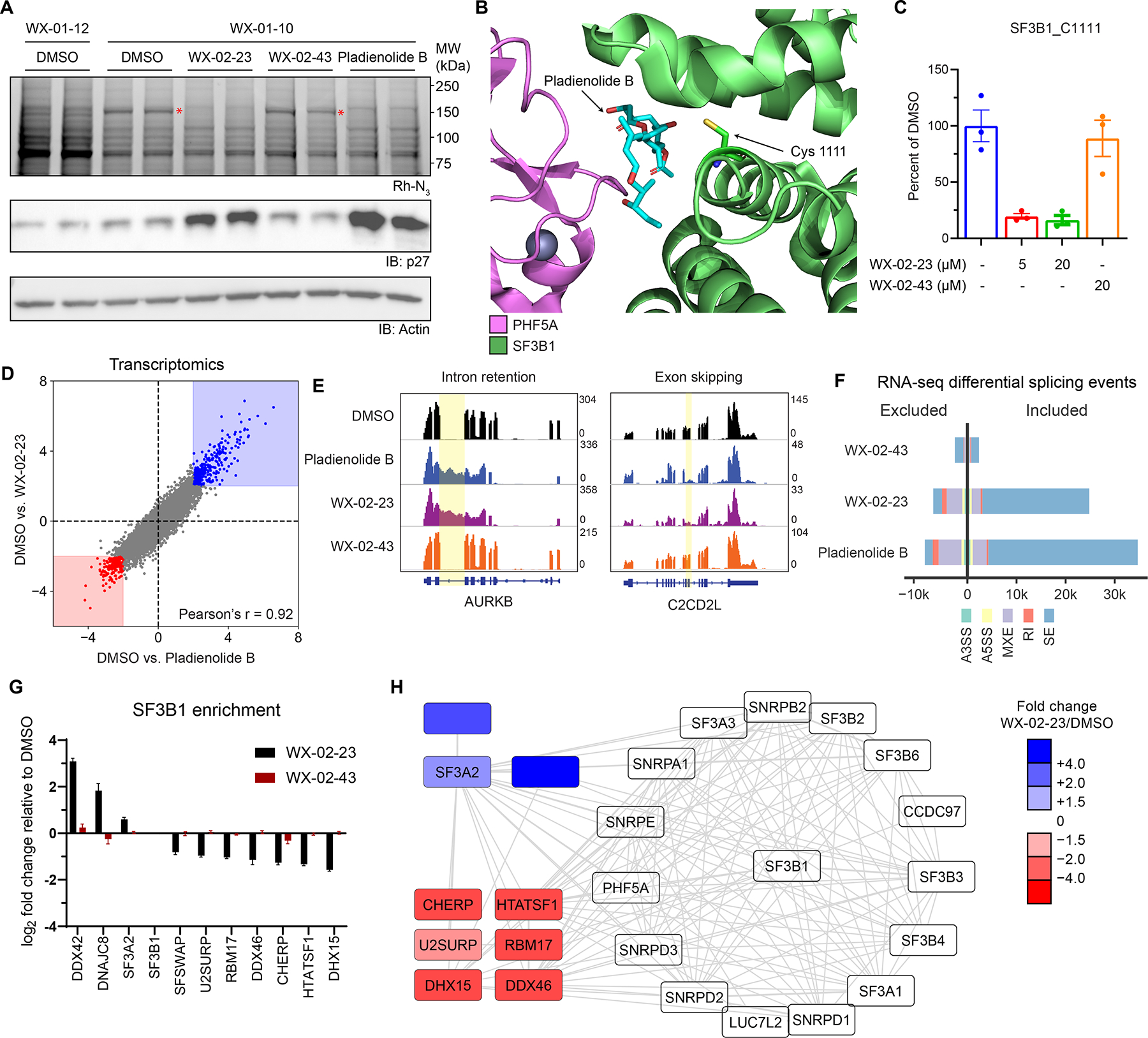
Tryptoline acrylamides engage C1111 of SF3B1 and stereoselectively modulate spliceosome structure and function. A. Stereoselective labeling of a 150 kDa protein (red asterisk) in 22Rv1 cells as determined by gel-ABPP. Top panel, Gel-ABPP, where cells were pre-treated with DMSO, WX-02-23 (1 μM), WX-02-43 (1 μM) or pladienolide B (10 nM) for 24 h followed by treatment with WX-01-10 or WX-01-12 (1 μM, 1 h). Lower panels, western blots. Results are from a single experiment representative of two independent experiments. B. Crystal structure of SF3B1-PHF5A complex bound to pladienolide B, highlighting the location of C1111 (PDB: 6EN4).
C. Quantification of stereoselective engagement of SF3B1_C1111 by WX-02-23 as measured by targeted cysteine-directed ABPP of 22Rv1 cells treated with 5 or 20 μM compound (3 h). Data are average values ± SD from n = 2–3 independent experiments.
D. Scatter plot of mRNA abundance changes in 22Rv1 cells treated with WX-02-23 (5 μM), pladienolide B (10 nM), or DMSO for 8 h. RNA-seq data are average values shown as log2 fold change relative to DMSO for n = 3 independent experiments.
E. Examples of intron retention in AURKB and exon skipping in C2CD2L caused by pladienolide B and WX-02-23 in 22Rv1 cells.
F. Summary of alternative splicing events caused by pladienolide B (10 nM), WX-02-23 (5 μM), and inactive enantiomer WX-02-43 (5 μM) in 22Rv1 cells (8 h) compared to DMSO treatment, as identified with rMATS by threshold of |PSI| > 0.1 and FDR < 0.05. Data represent values from three independent experiments.
G. Differential co-immunoprecipitation of proteins with SF3B1 (> 1.5 fold increase or decrease) in HEK293T cells treated with DMSO, WX-02-23, or WX-02-43 (5 μM, 3 h). Co-immunoprecipitated performed with anti-SF3B1 antibody (CST #14434). Data are average log2 fold changes ± SD from n = 4–7 independent experiments. See Dataset S1 for co-immunoprecipitation data from cells treated with pladienolide B (10 nM, 3 h).
H. Interactome map from STRING database filtered for proteins identified as SF3B1 interactors in the co-immunoprecipitation experiments from panel I. Data are average log2 fold changes from n = 4-7 independent experiments.
