Abstract
As the most abundant liver-specific microRNA, microRNA-122 (miR122) played a crucial role in the differentiation of stem cells into hepatocytes. However, highly efficient miR122 delivery still confronts challenges including poor cellular uptake and easy biodegradation. Herein, we for the first time demonstrated that the tetrahedral DNA (TDN) nanoplatform had great potential in inducing the differentiation of human mesenchymal stem cells (hMSCs) into functional hepatocyte-like cells (HLCs) by transferring the liver-specific miR122 to hMSCs efficiently without any extrinsic factors. As compared with miR122, miR122-functionalized TDN (TDN-miR122) could significantly up-regulate the protein expression levels of mature hepatocyte markers and hepatocyte-specific marker genes in hMSCs, indicating that TDN-miR122 could particularly activate the hepatocyte-specific properties of hMSCs for developing cell-based therapies in vitro. The transcriptomic analysis further indicated the potential mechanism that TDN-miR122 assisted hMSCs differentiated into functional HLCs. The TDN-miR122-hMSCs exhibited hepatic cell morphology phenotype, significantly up-regulated specific hepatocyte genes and hepatic biofunctions in comparison with the undifferentiated MSCs. Preclinical in vivo transplantation appeared that TDN-miR122-hMSCs in combination with or without TDN could efficiently rescue acute liver failure injury through hepatocyte function supplement, anti-apoptosis, cellular proliferation promotion, and anti-inflammatory. Collectively, our findings may provide a new and facile approach for hepatic differentiation of hMSCs for acute liver failure therapy. Further large animal model explorations are needed to study their potential in clinical translation in the future.
Keywords: Hepatocyte differentiation, Tetrahedral framework nucleic acids, Cell therapy, Transcriptomics, Acute liver failure treatment
Graphical abstract
Highlights
-
•
DNA tetrahedral nanomaterials are involved in the hepatic differentiation of hMSCs for the first time.
-
•
We establish a one-step approach to induce hepatic differentiation of hMSCs using miR122-functionalized tetrahedral DNA.
-
•
Cell therapy based on differentiated hepatocyte-like cells exhibits excellent therapeutic efficacy in acute liver failure.
1. Introduction
Acute liver failure (ALF) is a devastating and complicated hepatic disease caused by various endogenous or exogenous etiologies, such as autoimmunity or hereditary diseases, drug abuse, viruses, and toxins [1,2]. ALF is characterized by extensive hepatocellular necrosis, rapid onset of severe liver dysfunction, and accompanied by systemic inflammation and multiorgan failure, eventually leading to poor prognosis and high mortality [[3], [4], [5], [6]]. Until now, orthotopic liver transplantation is the only definitive curative treatment for ALF. Hence, the establishment of alternative therapeutic options for effectively treating ALF is still urgently needed due to the serious shortage of liver organ donors.
Cell therapy is currently an active research area for ALF treatment by direct injection of functional cells, such as hepatocytes and stem cells [[7], [8], [9]]. Although human hepatocytes have been well proven to improve liver function in both animal models and clinical trials with liver failure, obtaining adequate and functional hepatocytes remains a bottleneck for hepatocyte engraftment because of restricted availability and phenotypic instability [10]. As another candidate for cell therapy, stem cells with various subtypes have been extensively studied for ALF treatment, among which human mesenchymal stem cells (hMSCs) offer great therapeutic potential for ALF because of their easy availability and low immunogenicity [11,12]. Recent research has evidenced that hMSCs are able to differentiate into hepatocyte-like cells (HLCs) with hepatic-specific functions, which exhibit promising potential acting as the alternative cell source for ALF therapy [13,14]. However, the high cost and complicated differentiation procedures with customized cytokine cocktails may still limit its large-scale production [15].
As the most abundant liver-specific microRNA, microRNA-122 (miR122) played a crucial role in various physiological processes of liver development, including the differentiation of stem cells into hepatocytes [16,17]. Overexpression of miR122 can efficiently promote hepatic differentiation and maturation of stem cells through a miR122/FoxA1/HNF4a-positive feedback loop [18,19]. Despite the great advantage of miR122 in inducing hepatic differentiation of stem cells for ALF treatment, highly efficient miRNA delivery still confronts challenges such as instability, poor cellular uptake and easy biodegradation [20,21]. Recently, various organic and inorganic nanovehicles have been intensively studied for miRNA delivery, however, the inadequate delivery efficiency and safety concerns including potential cumulative toxicity and immunogenicity may limit their widespread applications [22]. Therefore, there is a highly urgent demand for miRNA delivery nanosystems with excellent delivery efficiency and high biocompatibility. The nucleic acid-based three-dimensional tetrahedral DNA nanostructures (TDNs) have attractive merits involving unsurpassed programmability, easy fabrication and good biocompatibility, which show promising potential as an alternative vehicle for the delivery of miRNA [[23], [24], [25], [26], [27], [28], [29]]. TDNs can not only facilitate the loading of negatively charged miRNA by direct extension or sticky-ended cohesion using complementary sequences [30,31], but are also able to directly enter cells by endocytosis without transfection agents based on the typical spatial structure [32,33]. In particular, TDNs have good biocompatibility and can protect miRNA from degradation by various enzymes within cells, which can enhance miRNA stability and improve cellular uptake and persistent response [[34], [35], [36]]. Therefore, TDNs are ideal for delivering miRNA to induce stem cell differentiation. Additionally, previous studies have shown that TDNs perform the capability to promote cell proliferation, migration, and angiogenesis, as well as enhance anti-inflammatory and antioxidant effects [[37], [38], [39]]. These unique biological activities make TDNs the appealing multifunctional miRNA nanocarrier for inducing stem cell hepatic differentiation in the synergistic treatment of liver failure.
In this study, we established an efficient TDNs-based miR122 gene delivery nanoplatform (TDN-miR122), which was sufficient to induce the human adipose mesenchymal stem cells (hMSCs) hepatic differentiation without additional cytokine support (Fig. 1). The TDN was self-assembled by mixing stoichiometric quantities of strands S1–S6, and miR122 was attached to TDN with the ratio of 6:1 via the hybridization to the single-stranded protruding sequences on the six side arms of TDNs to obtain the TDN-miR122. Notably, the TDN-miR122 could efficiently enhance the cellular uptake of miR122 and promote the differentiation of hMSCs to definitive functionally mature hepatocyte-like cells (TDN-miR122-hMSCs). Furthermore, the underlying action mechanism of TDN-miR122-induced mature hepatocyte-like cells was explored by RNA sequencing analysis. Additionally, after transplantation of the TDN-miR122-hMSCs into mice with carbon tetrachloride (CCl4)-induced ALF, we found that TDN-miR122-hMSCs could improve liver function and restore injured livers. Compared to hMSCs, the TDN-miR122-hMSCs exhibited greater characteristics in a lot of aspects of ALF therapy, such as the hepatocyte function supplement, anti-apoptosis, cellular proliferation promotion, and anti-inflammatory. In vivo results indicated that TDN-miR122-hMSCs alone, or in combination with TDN showed excellent therapeutic potential for CCl4-induced ALF. We expect that the TDNs-mediated miR122 delivery may serve as an effective approach to induce the differentiation of hMSCs into functional hepatocyte-like cells, which can provide a feasible cell source for hepatic regeneration for alleviating ALF injury. In addition, this simple and low-cost strategy for hMSCs hepatic differentiation may offer an alternative method of large quantities of hepatocytes production for clinical applications.
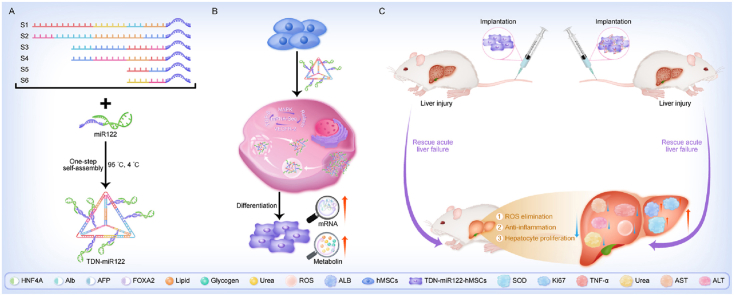
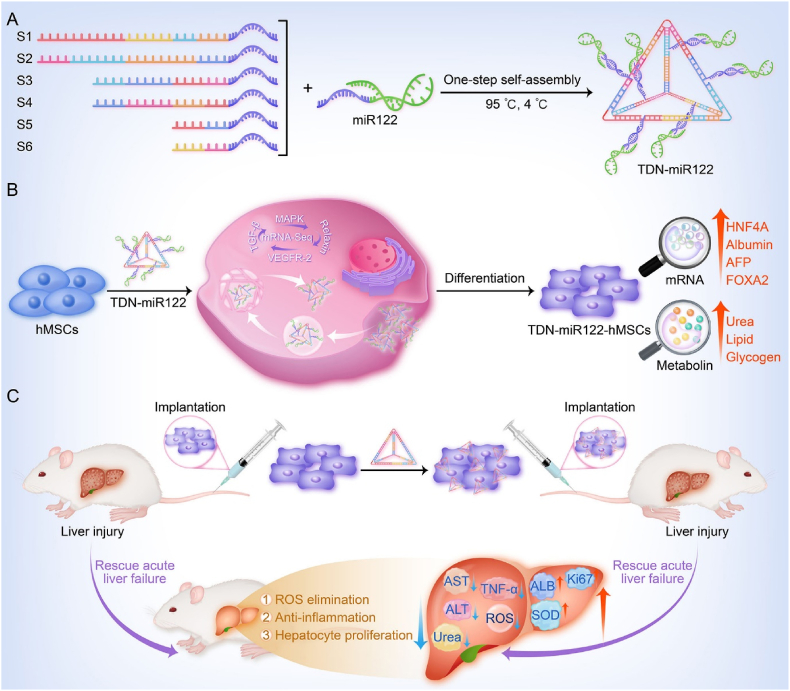
Fig. 1.
Schematic overview of TDN-miR122-hMSCs differentiated from hMSCs for the treatment of ALF. The TDN carrying miR122 can effectively enhance miR122 transfection and accelerate hepatic differentiation. The generated TDN-miR122-hMSCs exhibit mature hepatocyte functions and are capable of alleviating ALF injury. Especially, TDN not only serves as a carrier for inducing hMSCs hepatic differentiation, but also synergistically treats ALF.
2. Materials and methods
2.1. Fabrication and characterization of TDN-miR122
The formation of TDN-miR122 was carried out as follows: stoichiometric mixtures of component oligonucleotides (single-stranded DNA and double-stranded miRNA) were dissolved in annealing Tris-magnesium sulphate (TM) buffer solution (consisting of 10 mM Tris, 5 mM MgCl2, pH 8.0, Beyotime, China), heated to 95 °C for 5 min, then gently cooled to room temperature, and then incubated at 4 °C. The formations of the building unit and TDN-miR122 were characterized by agarose gel electrophoresis at a constant voltage of 120 V for 40 min. The agarose gel was immersed in an electrophoretic buffer containing ethidium bromide (EB, Sigma-Aldrich, USA) dye (0.5 μg/mL) for 20 min, and was scanned by a UV illumination (312 nm). The morphological features of TDN-miR122 were characterized by the atomic force microscopy (AFM, Nanoscope V multimode atomic force microscope (Dimension Icon (Bruker AXS, US))) image. The average hydrodynamic diameter of the nanoassembly was assessed by dynamic light scattering (DLS, Malvern Zetasizer (Nano ZS, Malvern, UK)).
2.2. In vitro biocompatibility evaluation
The human adipose tissue-derived mesenchymal stem cells (hMSCs) (2 × 104 cells/well) were plated in 96-well plates in 100 μL serum-free special medium (Saliai Stem cell Science and Technology Co, LTD, Guangzhou, China) and incubated overnight. Then the cells were incubated with TDN or TDN-miR122 (10 nM, 100 nM, 200 nM) for 24 h. Subsequently, the culture medium was replaced with cell counting kit-8 (CCK8) reagent for 2 h of incubation at 37 °C. The spectra microplate reader (BioTek, Synergy H1) was used to measure the absorbance at 450 nm. The untreated cells were used as the control group, only the medium was blank. Cell viability was determined as the following formula: Cell cytotoxicity (%) = (Asample - Ablank)/(Acontrol - Ablank) × 100.
2.3. Intracellular uptake of TDN-miR122
The hMSCs were separately seeded into 6-well plate and grown to around 90% confluency for 24 h before the experiments. After three times washing with phosphate buffer solution (PBS), the cells were then incubated with the culture medium containing Fluorescein (FAM)-labeled-TDN and FAM-labeled-TDN-miR122 for 2 h. After incubation, cells were collected and centrifuged at 1000 rpm for 5 min and washed twice with PBS buffer. Finally, the intracellular fluorescence of FAM-labeled-TDN-miR122 was quantified using a flow cytometer (NovoCyte, Santiago, California, USA). Fluorescence was determined by counting 20,000 events.
2.4. Subcellular locations and endocytosis mechanism
A colocalization assay was used to study the intracellular trafficking and localization of TDN-miR122 inside living cells. The hMSCs were seeded in a 35 mm confocal dish (coverglass-bottom dish) (MatTek Corp, Ashland, MA, USA) at a density of 1 × 104 cells/cm2 under standard culture conditions. Then, the cells were washed twice with PBS, and incubated with FAM-labeled-TDN-miR122 for 6 and 12 h. After that, the cells were washed by PBS 3 times and stained with 10 μM LysoTracker Red for 30 min and Hoechst 33342 for 15 min, respectively. Finally, the cells were washed with PBS buffer 3 times before being observed with a confocal laser scanning microscope (FV1000, Olympus, Tokyo, Japan). The excitation wavelengths were 405 nm and 543 nm, and the corresponding emission wavelengths were in the range of 430/480 nm and 550/630 nm for FAM and Hoechst 33342, respectively.
To investigate the endocytosis mechanism, cells (1 × 105) were pre-incubated with endocytosis inhibitors such as chlorpromazine (CPZ, 10 μM) or ethylisopropylamiloride (EIPA, 50 μM) before the addition of FAM-labeled-TDN/miR-122. After 30 min, FAM-labeled-TDN-miR122 (200 nM) was added to each sample in serum-free media and incubated at 37 °C in a CO2 incubator for 6 h. Cells were then harvested, washed twice with PBS (1 mL), and resuspended in ice-cold PBS (500 μL). The internalized TDN-miR122 was analyzed by a flow cytometer (Beckman, CytoFLEX, USA). For flow cytometric analysis of all samples, 10,000 events were recorded in triplicates and the internalization level was displayed with mean ± standard deviation (SD).
2.5. Hepatic differentiation and characterization
Human adipose tissue-derived mesenchymal stem cells (hMSCs) were cultured in the serum-free special medium (Saliai Stem cell Science and Technology Co, LTD, Guangzhou, China). The cells were grown at 37 °C humidified atmosphere containing 5% CO2. The cells were then seeded on 24-well culture plates at a density of 2 × 105 cells/mL. The cells were cultured and maintained in a serum-free special medium supplemented with TDN-miR122 (200 nM) for 21 days. The medium was changed every 2 days. In addition to morphological studies, we performed reverse-transcription polymerase chain reaction (RT-PCR) and transcriptome analyses of the in vitro hepatic differentiation of hMSCs to substantiate the efficiency of the differentiation process. Upon differentiation, hMSCs-derived hepatocyte-like cells (TDN-miR122-hMSCs) expressed several markers (Hepatocyte Nuclear Factor 4 Alpha (HNF4A), Forkhead box A2 (FOXA2), Albumin (ALB), Alpha fetoprotein (AFP)) in mature hepatocytes. The immunofluorescent staining shows that the TDN-miR122-hMSCs were positive for ALB, E-Cadherin (ECAD), and HNF4A and contained lipids and glycogen particles, all consistent with the results of differentiation to functional hepatocytes.
2.6. Quantitative real-time qRT-PCR for mRNA quantification
The different groups of cells were seeded on 6-well culture plates at a density of 1 × 106 cells/well. The total RNA was extracted from different groups of cells, according to the manufacturer's instructions for the RNA-Solv reagent. qRT-PCR was performed with standard HiScript II One Step qRT-PCR SYBR® Green Kit protocol. All samples were done in quadruplicate and normalized by the housekeeping gene beta-actin (β-actin). The RNA quantity and quality were evaluated by NanoDrop ND-2000 spectrophotometer (NanoDrop, Wilmington, DE, USA). The relative mRNA fold change was normalized to β-actin expression. The qRT-PCR primers are listed in Table S2.
2.7. Calcein-AM/Propidium Iodide (PI) double staining
Calcein-AM (Aladdin) stock solution was 1 mM calcein-AM in dimethyl sulfoxide (DMSO). The stock solution was diluted to 1–50 μM with PBS. The stock solution of PI (Solarbio) was diluted with PBS to 10–50 μM. The differentiating hMSCs were incubated with the calcein-AM and PI working solutions for 10–15 min. The samples were examined using an Ultra VIEW Vo X with excitation wavelengths of 488 and 561 nm.
2.8. Immunofluorescent staining
Cells were fixed with 4% paraformaldehyde (PFA) in PBS for 20 min at room temperature, then remove the PFA and washed with PBS. Next, the cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature. Following washing with PBS, the goat serum was blocked for 30 min and then washed twice with PBS. Cells were then incubated overnight with phalloidin-Alexa 488 (Invitrogen) and primary antibodies against ECAD (1:1000, Cell Signaling), albumin (1:1000, Invitrogen), hepatocyte nuclear factor 4α (HNF4α) (1:50, Cell Signaling) at 4 °C. Finally, the cells were washed with PBS and incubated for 2 h with secondary antibodies Alexa 488 Goat-anti-rabbit (1:500 in blocking solution; Invitrogen). Following washing with PBS, stained cells were imaged in confocal (Zeiss LSM 710) mode. 4′,6-diamidino-2-phenylindole (DAPI) was used to stain the nucleus for 5 min at room temperature.
2.9. In vivo fluorescence imaging
Briefly, the DiD labeled TDN-miR122-hMSCs were administered into ALF mice via tail intravenously injection. The mice were imaged with excitation at 640 nm and emission at 700 nm after 3 days. The noninvasive NIR fluorescence imaging system (IVIS Lumina LT Series III, PerkinElmer, USA) consists of an excitation light source and a cold charge coupled device (CCD).
2.10. RNA extraction and library construction
Total RNA was isolated and purified using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's procedure. The RNA amount and purity of each sample were quantified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). The RNA integrity was assessed by Bioanalyzer 2100 (Agilent, CA, USA) with RIN number >7.0, and confirmed by electrophoresis with denaturing agarose gel. Poly (A) RNA is purified from 1 μg total RNA using Dynabeads Oligo (dT) 25–61005 (Thermo Fisher, CA, USA) using two rounds of purification. Then the poly(A) RNA was fragmented into small pieces using Magnesium RNA Fragmentation Module (NEB, cat.e6150, USA) under 94 °C for 5–7min. Then the cleaved RNA fragments were reverse-transcribed to create the cDNA by SuperScript™ II Reverse Transcriptase (Invitrogen, cat. 1896649, USA), which were next used to synthesize U-labeled second-stranded DNAs with E. coli DNA polymerase I (NEB, cat.m0209, USA), RNase H (NEB, cat.m0297, USA) and dUTP solution (Thermo Fisher, cat.R0133, USA). An A-base is then added to the blunt ends of each strand, preparing them for ligation to the indexed adapters. Each adapter contains a T-base overhang for ligating the adapter to the A-tailed fragmented DNA. Single- or dual-index adapters are ligated to the fragments, and size selection was performed with AMPureXP beads. After the heat-labile UDG enzyme (NEB, cat.m0280, USA) treatment of the U-labeled second-stranded DNAs, the ligated products are amplified with PCR by the following conditions: initial denaturation at 95 °C for 3 min; 8 cycles of denaturation at 98 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 30 s, and then final extension at 72 °C for 5 min. The average insert size for the final cDNA library was 300 ± 50 bp. At last, we performed the 2 × 150 bp paired-end sequencing (PE150) on an illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China) following the vendor's recommended protocol.
2.11. Sequence and primary analysis
Cutadapt software (https://cutadapt.readthedocs.io/en/stable/,version:cutadapt-1.9) was used to remove the reads that contained adaptor contamination (command line: ∼cutadapt -a ADAPT1 -A ADAPT2 -o out1.fastq -p out2.fastq in1.fastq in2.fastq -O 5 -m 100). After removing the low-quality bases and undetermined bases, we used HISAT2 software (https://daehwankimlab.github.io/hisat2/,version:hisat2-2.0.4) to map reads to the genome (for example:Homo sapiens Ensembl v96), (command line: ∼hisat2 -1 R1.fastq.gz −2 R1.fastq.gz -S sample_mapped.sam). The mapped reads of each sample were assembled using StringTie (http://ccb.jhu.edu/software/stringtie/, version:stringtie-1.3.4d.Linux_x86_64) with default parameters (command line: ∼ stringtie -p 4 -G genome.gtf -o output.gtf -l sample input.bam). Then, all transcriptomes from all samples were merged to reconstruct a comprehensive transcriptome using gffcompare software (http://ccb.jhu.edu/software/stringtie/gffcompare. shtml, version: gffcompare - 0.9.8. Linux x86_64). After the final transcriptome was generated, StringTie and ballgown (http://www.bioconductor.org/packages/release/bioc/html/ballgown.html) were used to estimate the expression levels of all transcripts and perform expression level for mRNAs by calculating FPKM (FPKM = [total_exon_fragments/mapped_reads(millions) × exon _length (kB)]), (command line: ∼stringtie -e -B -p 4 -G merged.gtf -o samples.gtf samples. bam). The differentially expressed mRNAs were selected with fold change >2 or fold change <0.5 and p value < 0.05 by R package edgeR (https://bioconductor.org/packages/release/bioc/html/edgeR.html) or DESeq2 (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html), and then analysis GO enrichment and KEGG enrichment to the differentially expressed mRNAs.
2.12. Transplantation of TDN-miR-122-hMSCs into mice with ALF
The 5-week-old male C57BL/6J mice were housed in laminar flow cabinets. The food, water, bedding, and cages were all autoclaved before use. As described in previous studies, the mice were intraperitoneally injected with 4 μL/g (mouse weight) CCl4 in an olive oil solution to induce ALF [3]. ALF was confirmed by liver histology and blood biochemistry 24 h after CCl4 injections. All mice were then randomly divided into eight groups (fifteen mice for each group). The first group was untreated male mice as a negative control group. 24 h after the model induction, groups 2–8 were intravenously injected with 100 μL 0.9% saline as a negative control, hMSCs (5 × 106), TDN-hMSCs (5 × 106), miR-122-hMSCs (5 × 106), TDN-miR-122-hMSCs (5 × 106), respectively. The CCl4-induced ALF mice with or without treatment with TDN-miR122-hMSCs were assessed after 3 days of cell transplantation. Finally, mice were euthanized and the main organ sections were collected and fixed in the 10% neutral-buffered formalin and their liver and kidney functions were compared with that of healthy control mice. To assess the curative effect of the TDN-miR122-hMSCs on the liver structures and functions, the serum was collected to test the alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels using an ALT/AST assay kit. For the histological analysis, the dissected liver was weighed and then divided into three parts: one for H&E staining and BrdU and nuclear protein Ki67 immunohistochemical staining and the other two for reactive oxygen species (ROS) and tumor necrosis factor-α (TNF-α) analyses. The study protocol was reviewed and approved by the Ethical Review Committee for Laboratory Animal Welfare of Sun Yat-sen University. Written informed consent was obtained from all participants. All experiments were performed in accordance with relevant guidelines and regulations.
2.13. Histological and serological evaluation
To evaluate the therapeutic effect and biosafety of TDN-miR122-hMSCs, ALF mice were euthanized, and the major organs such as heart, liver, spleen, lung and kidney were extracted and fixed in 10% buffered formalin. The tissues were embedded in liquid paraffin and then were sectioned into 4 μm thick slices. Slices were stained with hematoxylin and eosin (H&E). The Images were evaluated using a standard upright histology microscope (Olympus DX45, Japan) for notable histological observation and histopathological analysis. For frozen tissue staining, 10 μm thick liver cryosections were fixed with 4% paraformaldehyde and permeabilized/blocked. To identify proliferating cells in liver tissue, the slides were incubated with anti-Ki-67 (1:100) and counterstained with DAPI which was used for nuclei staining. Besides, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining and counterstaining with DAPI were used in the assessment of cell apoptosis. Images were taken with a confocal microscope. Finally, superoxide dismutase (SOD) levels in liver homogenate were determined. To evaluate the therapeutic efficacy, liver-related ALT and AST, as well as kidney-associated serum urea nitrogen (UN) and serum creatinine (CREAT) toxicity were measured. Moreover, the main cytokine (TNF-α) as a systemic inflammation marker was analyzed.
2.14. Statistical analysis
All statistical data were analyzed by GraphPad Prism 6.0 (GraphPad Software). Results are represented as mean ± standard deviation (SD). A one-way analysis of variance (ANOVA) test and Two-tailed Student's t-test or 2-sided Welch's t-test were used as appropriate. To ensure statistical power, the numbers of experimental replicates are at least three replicates in vitro, the mice experimental groups were typically composed of 5 animals each. The statistical significance is represented by the bars and asterisks where *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns is not significant.
Ethical statement
All experimental protocols using hMSCs primary cells, and animal experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) and Animal Experiment Center of Sun Yat-sen University. All animals were cared for in accordance with the requirements of the Laboratory Animal Welfare Act and amendments thereof.
3. Results and discussion
3.1. Characterization of TDN-miR122
The TDN-miR122 was prepared through a careful annealing process by one-step self-assembly of tetrahedral skeleton DNA fragments and miR122 (Table S1) at the ratio of 1:6 based on the DNA self-assembling design principle and theoretical evaluation of UNPACK and SEQUIN programs [40]. The agarose gel electrophoresis analysis demonstrated the stepwise assembly of TDN as each strand was added (Fig. 2A). The resultant DNA nanoassembly showed a single band, indicating that the TDN-miR122 was successfully prepared with a high yield. The atomic force microscopy (AFM) images revealed that TDN was nanosized particles with a diameter of about 55 nm (Fig. 2B). After miR122 modification, the size of TDN-miR122 increased slightly to about 65 nm (Fig. 2C). Dynamic light scattering (DLS) and zeta potential characterizations were further applied to confirm the conjugation of miR122 to TDN. As shown in Fig. 2D and E, the TDN had an average size of 58.81 ± 3.63 nm and a zeta potential of −13.37 ± 0.54 mV. After miR122 loading, the average size of TDN-miR122 increased to 65.26 ± 4.34 nm and the zeta potential decreased to −25.89 ± 3.13 mV. The polydispersity indexes of TDNs and TDN-miR122 are 29.32% ± 1.67% and 27.51% ± 1.56%, respectively. Furthermore, although miR122 with 2′-OMe-modified bases could enhance serum stability, the TDN-miR122 showed further greatly improved serum stability compared with miR122 (Fig. S1), indicating that TDN-miR122 endowed miR122 cargo effective resistance against unfavorable degradation and sustained release under physiological conditions.
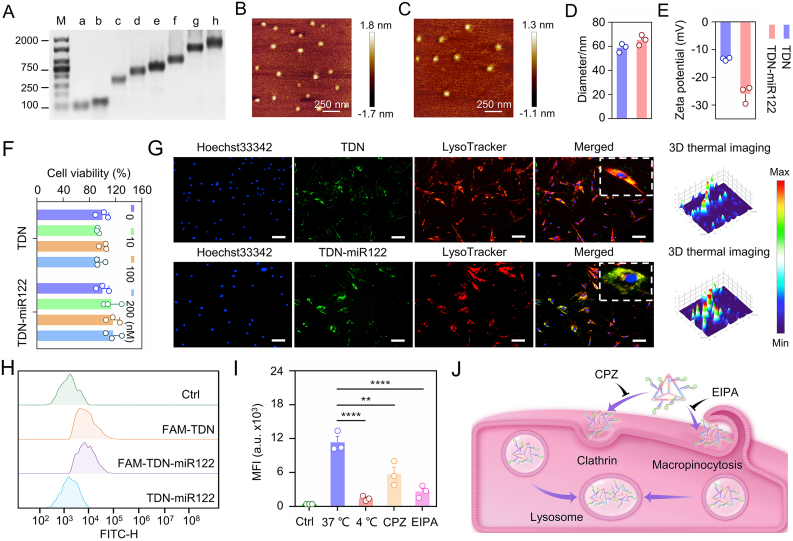
Fig. 2.
Characterizations of TDN and TDN-miR122. (A) Agarose gel electrophoresis showing the successful synthesis of the TDN-miR122 (M: Marker, a: miR-122, b: S1, c: S1+S2, d: S1+S2+S3, e: S1+S2+S3+S4, f: S1+S2+S3+S4+S5, g: S1+S2+S3+S4+S5+S6 (TDN), h: TDN-miR122). (B–C) The AFM images of the TDN (B) and TDN-miR122 (C). (D) Hydrodynamic diameters of the TDN and TDN-miR122. (E) Zeta potentials of TDN and TDN-miR122. (F) The cytotoxicity of various concentrations of TDN and TDN-miR122 towards hMSCs at 24 h. (G) Fluorescent images of cellular uptake of TDN and TDN-miR122-FAM in hMSCs after incubation for 6 h (scale bar: 50 μm). Red: LysoTracker; Green: FAM-labeled TDN and TDN-miR122; blue: Hoechst33342-stained nuclei. (H) FACS evaluation to monitor the TDN and TDN-miR122 uptake in hMSCs for 2 h. (I) Cellular uptake efficiency of TDN-miR122-FAM in hMSCs in the presence of endocytosis inhibitors: chlorpromazine (CPZ, 10 μM, clathrin-mediated endocytosis), 5- (N-ethyl-N-isopropyl) amiloride (EIPA, 50 μM, macropinocytosis). The lowered uptake levels at 4 °C indicated endocytosis of TDN-miR122. Data are represented as mean ± standard deviation (SD) (n = 3). ****P < 0.0001; ***P < 0.001; **P < 0.01. (J) Schematic illustration of the cellular uptake pathways of TDN-miR122.
High biosafety is essential for using the TDN nanoplatform for miR122 delivery. The cytotoxicities of TDN and TDN-miR122 at different concentrations toward hMSCs were analyzed using the cell counting kit-8 (CCK8) kits. As shown in Fig. 2F, both TDN and TDN-miR122 exhibited minimal effects on cell viability, even at high concentrations, indicating the good biocompatibility of TDN-miR122. According to the CCK8 assays, 100 nM was determined as the optimal concentration of TDN and TDN-miR122 for subsequent studies. To evaluate the hepatic differentiation capability of TDN-miR122, we first assessed its capability to enter hMSCs, which was the first step before it could perform its biofunction to induce differentiation. As illustrated in Fig. 2G and H, the fluorescent images and flow cytometer (FACS) analysis revealed that TDNs carrying miR122 did not affect their entry into hMSCs. Furthermore, the TDN-miR122 could successfully escape from lysosomes and be released into the cytoplasm at 6 h, which was important for the bioapplications of miR122 in hMSCs. To further investigate the pathway mediating the cellular uptake of the TDN-miR122, the impacts of energy inhibition (4 °C), and two endocytosis inhibitors (chlorpromazine, an inhibitor of clathrin-mediated endocytosis, and 5-(N-ethyl-N-isopropyl) amiloride, an inhibitor of macropinocytosis) for cell entry were evaluated. The FACS analysis and the corresponding statistical results in Fig. 2I and S2 proved that the cellular uptake of TDN-miR122 was greatly suppressed by both temperature decrease and endocytosis-related inhibitors, suggesting that the cellular uptake of TDN-miR122 primarily followed the energy-dependent and endocytosis-dependent pathways (Fig. 2J).
3.2. TDN-miR122 inducing functional hepatocyte-like cells from hMSCs
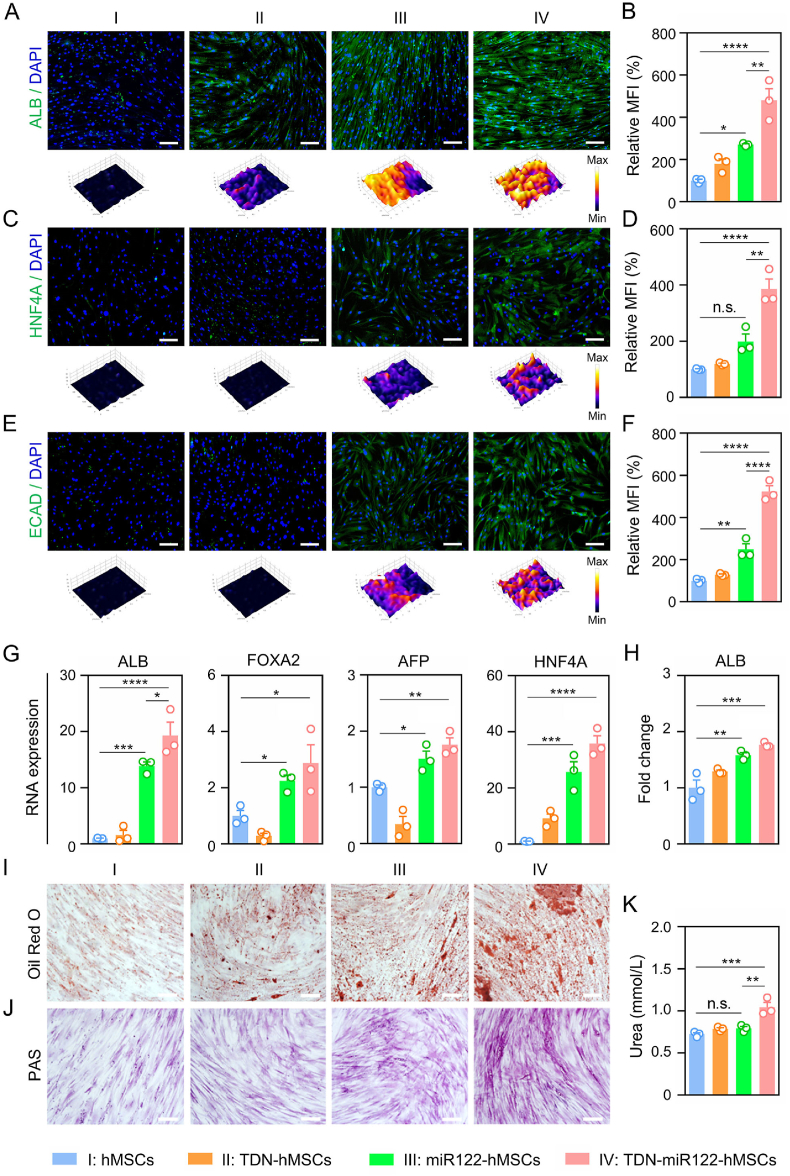
Afterwards, to determine the effects of TDN-miR122 during hepatic differentiation of hMSCs, the hMSCs were exposed to the cell culture medium in the presence or absence of TDN-miR122 over the course of 21 days. The remarkable morphological changes were observed after 21 days of differentiation induced by TDN-miR122 (Fig. S3). Undifferentiated hADSCs presented a typical fibroblast-like cell shape, while TDN-miR122-induced hepatocyte-like cells (TDN-miR122-hMSCs) exhibited the polygonal and round morphologies. To determine the maturation level and fate of the hepatocyte-like cells after TDN-miR122-induced differentiation for 21 days, the protein expression levels of mature hepatocyte markers in cells, such as albumin (ALB), hepatocyte nuclear factor 4 alpha (HNF4A) and E-cadherin (ECAD) were assessed. The immunofluorescent staining and the corresponding quantitative data (Fig. 3A, B) indicated that the significant up-regulation of ALB expression was observed in the TDN-miR122-hMSCs group as compared to the hMSCs group and the miR122-induced hepatocyte-like cells (miR122-hMSCs) group, which quantitatively increased 4.80-fold and 1.77-fold, respectively. Moreover, the levels of HNF4A in cells treated with TDN-miR122 were increased approximately 3.86-fold and 1.94-fold compared with those in untreated hMSCs and hMSCs treated with miR122, respectively (Fig. 3C, D). Similarly, ECAD expression levels in the TDN-miR122-hMSCs group were 5.23-fold and 2.01-fold higher than those in the hMSCs group and the miR122-hMSCs group (Fig. 3E, F), similar to expression levels in the human normal liver cell (LO2) (Fig. S4). The above phenomena suggested that TDN-miR122 could efficiently promote hepatic-directed differentiation as compared to miR122.
Fig. 3.
TDN-miR122 inducing functional hepatocyte-like cells from MSCs. (A-F) Immunofluorescent staining of hepatocyte markers, including ALB (A–B), HNF4A (C–D) and ECAD (E–F) expressions in the TDN-miR122 induced MSCs-Hep cells (TDN-miR122-MSCs) for 21 days, and the corresponding quantified results. Nuclei were stained with DAPI (scale bar: 100 μm). (G) The mRNA expression levels of hepatocyte-specific marker genes (ALB, FOXA2, HNF4A and AFP) in differentiated cells at 21 days. (H) The detection of ALB in the cell culture medium of TDN-miR122-hMSCs after 21 days. (I) The characteristics of mature hepatocytes. Lipid droplets in TDN-miR122-hMSCs at 21 days were stained red by Oil Red O and observed under the microscope (scale bar: 100 μm). (J) Abilities for glycogen synthesis in TDN-miR122-hMSCs. Photos of periodic acid-Schiff (PAS) stain in different groups for 21 days (scale bar: 100 μm). (K) The detection of urea in the cell culture medium of TDN-miR122-hMSCs after 21 days.
Subsequently, the expressions of hepatocyte-specific markers genes in differentiated cells at 21 days, including ALB, FOXA2, AFP, and HNF4A, were also characterized by quantitative real-time polymerase chain reaction (qRT-PCR) analysis (Fig. 3G). The gene expression of ALB was markedly upregulated in the hMSCs treated with TDN-miR122 compared with control hMSCs (19.30-fold) and miR122-treated MSCs (1.39-fold). In addition, the mRNA expressions of FOXA2, AFP, and HNF4A were also significantly upregulated. Furthermore, compared to hMSCs and miR122-hMSCs, the induced TDN-miR122-hMSCs exhibited much higher levels of lipid accumulation and glycogen production as evidenced by Oil Red O staining and periodic acid-Schiff (PAS) staining (Fig. 3I and J), representing important biofunctions of mature hepatocytes (Fig. S5). Additionally, it was found that the TDN-miR122-hMSCs could effectively enhance the ALB secretion (Fig. 3H) and urea production (Fig. 3K) in the medium supernatants. Taken together, all these results proved that TDN-miR122-induced hMSCs could be successfully differentiated into hepatocyte-like cells exhibiting typical mature hepatic functional features, which might be great candidates to improve liver biofunctions in ALF.
3.3. Transcriptome analysis reveals the underlying differentiation mechanism of TDN-miR122-hMSCs
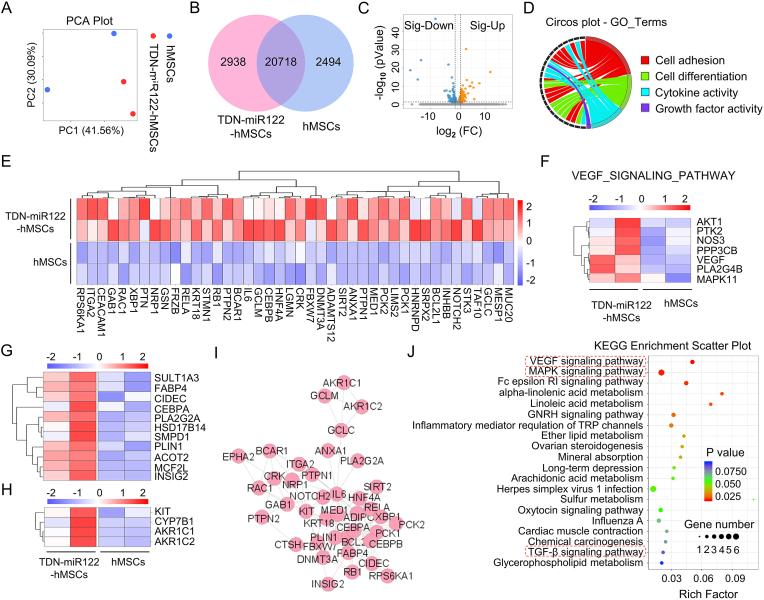
To further investigate the differentiation phenotype and the molecular profiles of the TDN-miR122-hMSCs generated with our protocol, we performed mRNA sequencing on hMSCs and TDN-miR122-hMSCs. As displayed in Fig. 4A, the principal component analysis (PCA) demonstrated that the expressed genes of the TDN-miR122-hMSCs group were extremely distinct from the hMSCs group. The venn diagram showed that 20718 genes were co-expressed by two groups, whereas 2938 genes were exclusively expressed in the TDN-miR122-hMSCs group (Fig. 4B). Volcano plots and heatmaps then further highlighted 224 differentially expressed genes (DEGs), of which 139 and 85 genes were upregulated and downregulated, respectively (Fig. 4C, S6). Moreover, Gene Ontology (GO) enrichment analysis showed that the DEGs were mainly enriched in 25 biological processes (Fig. S7, S8). Notably, cell adhesion, lipid metabolism, as well as multicellular organization development as the significantly enriched terms, would affect the developmental potential of liver organoids. In particular, the chord diagram of GO categories showed that the genes upregulated in TDN-miR122-hMSCs group were mainly enriched in "Cell differentiation", "Cell adhesion", "Cytokine activity" and "Growth factor activity", accounting for more than 50.0% of all the DEGs (Fig. 4D). To further characterize the hepatic differentiation and maturation of TDN-miR122-hMSCs compared with the hMSCs, the expression profiles of mature liver signature genes were analyzed. The heatmaps showed that the expressions of hepatocyte-related genes (Fig. 4E), genes related to bile acid synthesis (Fig. 4G), as well as the fat and lipid metabolism genes (Fig. 4H) were obviously upregulated in the TDN-miR122-hMSCs group.
Fig. 4.
Transcriptome analysis reveals the underlying action mechanism of TDN-miR122 induced hMSCs-Hep. (A) Principal component analysis (PCA) was performed based on differentially expressed genes from TDN-miR122-hMSCs and hMSCs groups. Each data point corresponds to the PCA analysis of each sample. (B) Venn diagram of mRNAs differentially expressed in TDN-miR122-hMSCs groups and hMSCs groups. (C) A volcano plot showing the differences in gene expression between TDN-miR122-hMSCs groups and hMSCs groups. Blue points represent genes that were significantly reduced with a fold change >2, and red points represented significantly upregulated genes with a fold change >2. Gray points represents no significant difference. (D) Gene ontology (GO) enrichment analysis of the subordination between the representative differentially expressed genes and their enriched pathways. (E) Heatmap representing the expression of genes relating to specific liver functions (hepatocyte-related genes) in TDN-miR122-hMSCs groups and hMSCs groups, which further verified that hMSCs-Hep had the biological function of hepatocytes. (F) Heatmap representing the expression of genes relating to the VEGF signaling pathway in TDN-miR122-hMSCs groups and hMSCs groups. (G) Heatmap representing the expression of genes relating to the bile acid synthesis in TDN-miR122-hMSCs groups and hMSCs groups. (H) Heatmap representing the expression of genes relating to the fat and lipid metabolism in TDN-miR122-hMSCs groups and hMSCs groups. (I) PPI network consists of representative genes related to hepatic function and metabolism process markers, circles represent genes, and area of the circle represents the degree of genes. (J) Bubble charts show the KEGG pathway analysis results of differentially expressed genes (Top 20 KEGG pathways of differentially expressed genes).
Moreover, we conducted a protein-protein interaction (PPI) network analysis of representative DEGs involved in the genes relating to specific liver functions using the STRING database (Fig. 4I), which revealed the functional linkages among proteins encoded by DEGs. To further illustrate the mechanism of action, the heatmap analysis figured out that TDN-miR122-hMSCs promoted the up-regulation of related genes in the VEGF signaling pathway, which was highly associated with the hepatocytes maturation of the TDN-miR122-hMSCs (Fig. 4F). Simultaneously, to determine the main affected pathways, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis indicated that the DEGs were remarkably enriched in TGF-β signaling pathway, MAPK signaling pathway, VEGF signaling pathway, and so on, which were highly associated with the liver differentiation and liver homeostasis (Fig. 4J). Interestingly, the KEGG analysis of the DEGs was consistent with our findings (Fig. 4F), which would be of great help to clarify the molecular mechanism underlying the conversion of hMSCs into hepatocyte-like cells.
3.4. In vivo therapeutic efficiency of TDN-miR122-hMSCs on ALF mice
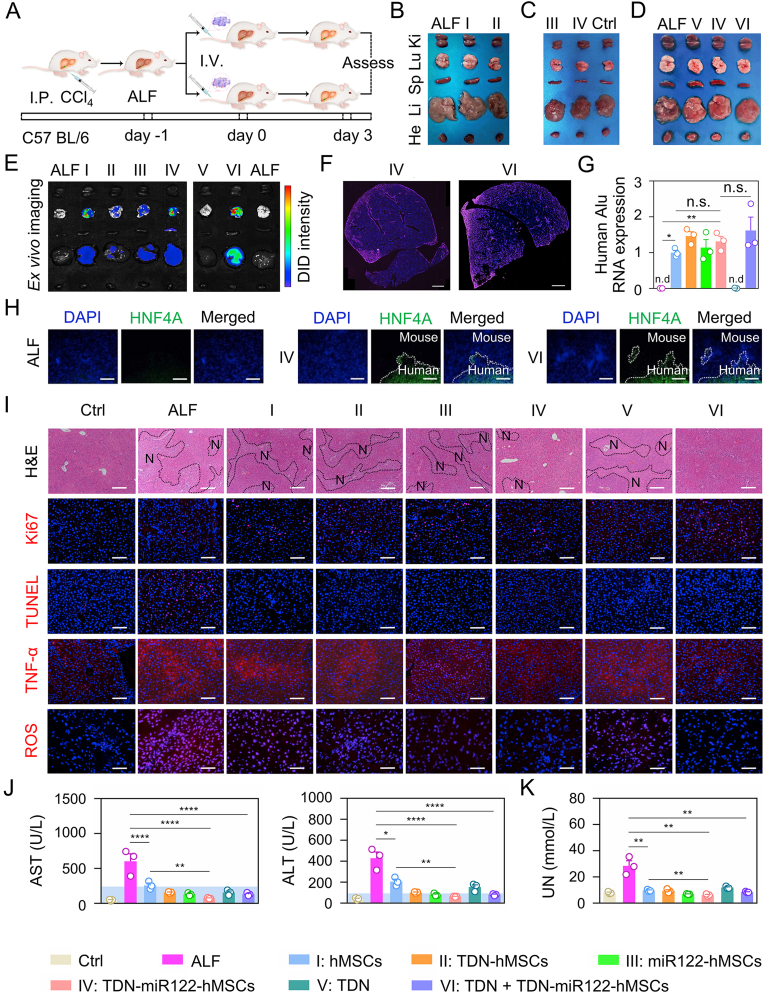
Encouraged by the impressive hepatic functional features of TDN-miR122-hMSCs, we then investigated the in vivo therapeutic performance of TDN-miR122-hMSCs in the CCl4-induced ALF model (Fig. 5A). Previous studies have indicated that intravenous injection is a common route for cell transplantation in both preclinical and clinical research due to its minimal invasion and easy operation [41,42]. However, the high ROS level and inflammatory environment in the ALF liver may cause apoptosis and necrosis of transplanted cells [5,43,44]. Therefore, we also used TDN as the protective nanoagent for injecting TDN-miR122-hMSCs since TDN possessed unique anti-inflammatory and antioxidant activities [34,45]. The therapeutic transplantation procedures were illustrated in Fig. 5A. On day 3 after treatment, the mice were sacrificed, and the primary organs (heart, liver, spleen, lung and kidney) were dissected. As shown in Fig. 5B–D, the TDN-miR122-hMSCs group showed a significant therapeutic effect, while the serious edematous pale liver morphologies were observed in the ALF (CCl4-pretreatment) group and hMSCs-treated group. Likewise, the livers in the TDN-miR122-hMSCs combined with the TDN treatment group presented a smooth surface with homogenous dark brown color, exhibiting no difference compared with the control (healthy mice) group.
Fig. 5.
In vivo therapeutic efficiency of TDN-miR122-hMSCs on ALF mice. (A) Schematic diagram of the experimental procedure. Intravenous injection (I.V.), Intraperitoneal injection (I.P.). (B–D) Digital photos of major organs from ALF mice after various treatments at the end of the experiment (I: undifferentiated hMSCs, II: TDN treated hMSCs, III: miR122 treated hMSCs, IV: TDN-miR122 treated hMSCs, V: free-TDN, VI: TDN + TDN-miR122 treated hMSCs). (E) In vivo fluorescence biodistribution images of various tissues (hearts, livers, spleens, lungs and kidneys) at 3 days after intravenous injection of the DiD labeled different cells. (F) The whole liver scanning distribution image of TDN-miR122-hMSCs group and TDN-miR122-hMSCs combined with TDN treatment group, cells were labeled with DiD dye (scale bar: 1 mm). (G) Quantitative real-time PCR (qRT-PCR) was used to measure human Alu-sequences specificity (the most conserved region in human DNA) for the identification and quantification of human cell lines (rose red: ALF, blue: hMSCs, orange: TDN treated hMSCs, green: miR122 treated hMSCs, pink: TDN-miR122 treated hMSCs, dark green: free-TDN, purple: TDN + TDN-miR122 treated hMSCs). (H) Immunofluorescent staining of hepatocyte marker human HNF4α expression in liver tissue of TDN-miR122-hMSCs groups and TDN-miR122-hMSCs with TDN added treatment group (scale bar: 100 μm). (I) H&E stain (N, necrosis), Ki67-positive cells (red nuclei) and TUNEL-positive cells (red nuclei), TNF-α and ROS concentrations (Scale bar: 50 μm) of the liver tissues from ALF mice treated with different groups on day 3 (Scale bar: 100 μm) (I: undifferentiated hMSCs, II: TDN treated hMSCs, III: miR122 treated hMSCs, IV: TDN-miR122 treated hMSCs, V: free-TDN, VI: TDN + TDN-miR122 treated hMSCs). (J) Analysis of hematological markers of liver damage (Normal range (blue background). ALT, alanine aminotransferase; AST, aspartate aminotransferase in the ALF mice after different treatments. (K) The detection of urea nitrogen (UN) in the serum of mice after various treatments.
To delineate the exact localization and growth pattern of the infused TDN-miR122-hMSCs, the recipients were sacrificed 3 days after transplantation and the presence of TDN-miR122-hMSCs in different internal organs was evaluated. As illustrated in Fig. 5E and S9, the labeled hMSCs and TDN-miR122-hMSCs were predominantly present in the lung and liver, and no substantial TDN-miR122-hMSCs migration into other organs was detected. It was worth noting that, as compared with other treatment groups, the co-injection of TDN-miR122-hMSCs and TDN improved the TDN-miR122-hMSCs accumulation in the liver, implying that TDN could help reduce the TDN-miR122-hMSCs damages induced by oxidative stress and inflammation in ALF lesion [36,46,47], which further enhanced the utilization of TDN-miR122-hMSCs. In addition, the fluorescence scanning imaging results of the whole liver (Fig. 5F) showed that the strong fluorescence of TDN-miR122-hMSCs in liver tissues could be observed in both TDN-miR122-hMSCs group and TDN + TDN-miR122-hMSCs group, mainly accumulated around the blood vessels of the liver, further suggesting the transplanted TDN-miR122-hMSCs were capable of engrafting in the ALF mice liver.
To further investigate the homing level of TDN-miR122-hMSCs, we performed the qRT-PCR analysis of human-specific Alu sequences in ALF mice liver tissues among different treatment groups (Fig. 5G). The high Alu levels of the TDN-miR122-hMSCs group and the TDN + TDN-miR122-hMSCs group in ALF mouse liver indicated that TDN-miR122-hMSCs maintained good migration and growth ability in ALF mice. Furthermore, immunofluorescence staining also helped distinguish the existence of human hepatocytes in the liver tissue of ALF mice via the human species-specific markers of HNF4α after the TDN-miR122-hMSCs and the TDN + TDN-miR122-hMSCs treatments (Fig. 5H). Overall, all these results indicated that the TDN-miR122-hMSCs grafts, combined with or without TDN treatment, exhibited great therapeutic potential for ALF in vivo.
After demonstrating that TDN-miR122-hMSCs could be efficiently engrafted into the liver of ALF mice, we then evaluated the in vivo therapeutic performance of TDN-miR122-hMSCs. As shown in Fig. 5J, the serum levels of AST and ALT in mice, which were typical diagnostic indicators for ALF, decreased significantly on day 3 after the intravenous injection of hADMSCs and TDN-miR122-hMSCs. Particularly, the AST and ALT levels in the TDN-miR122-hMSCs treatment group tended to return to normal levels. Interestingly, the injection of TDN through tail vein also had a certain therapeutic potential. Notably, TDN significantly increased the SOD level in the ALF liver (Fig. S10). Additionally, the levels of serum urea nitrogen (UN) and serum creatinine (CREAT) decreased significantly (Fig. 5K and S11), indicating that the TDN-miR122-hMSCs treatment groups avoided the complications caused by ALF, such as renal failure.
To further evaluate the therapeutic effectiveness and safety, the biochemical indices were assessed from liver tissue sections. As illustrated in Fig. 5I, livers in the ALF, hMSCs, TDN-hMSCs, miR122-hMSCs and TDN groups showed massive liver necrotic damage, while the livers in the TDN-miR122-hMSCs group and the TDN + TDN-miR122-hMSCs group displayed minimal necrosis, proving the efficient therapeutic capacity of TDN-miR122-hMSCs. In addition, no significant morphological and histopathological abnormalities were observed in major organs (heart, spleen, lung, and kidney) stained with the hematoxylin and eosin (H&E) in the TDN-miR122-hMSCs group and the TDN + TDN-miR122-hMSCs group (Fig. S12), demonstrating that TDN-miR122-hMSCs and TDN had no serious side effects and systemic toxicity. Ki67 staining and TUNEL immunofluorescence analysis revealed that the TDN-miR122-hMSCs group and the TDN + TDN-miR122-hMSCs group could effectively promote hepatocyte proliferation and inhibit apoptosis, indicating a high level of liver protection and regeneration (Fig. 5I). For further validating the effect of alleviating oxidative stress and inflammation of TDN-miR122-hMSCs, we assessed the levels of TNF-α and ROS in mice livers. TNF-α immunofluorescence analysis and ROS immunofluorescence analysis exhibited that the TDN + TDN-miR122-hMSCs treatment highly improved the antioxidant and anti-inflammatory levels in liver tissue. Collectively, all these findings suggested that the TDN + TDN-miR122-hMSCs treatment could efficiently scavenge excessive ROS, suppress the inflammatory response, reduce hepatocellular necrosis, and eventually improve hepatic regeneration, exhibiting excellent therapeutic efficacy in a mouse ALF model.
4. Conclusion
In summary, we fabricated the TDNs-based miR122 gene delivery nanoplatform to induce the hepatic differentiation of hMSCs to functionally mature hepatocyte-like cells for ALF therapy. On the one hand, the TDN-miR122 could effectively improve the cellular uptake efficiency of miR122 through macropinocytosis and caveolae-mediated endocytosis. On the other hand, TDN-miR122 could efficiently promote the hepatic-directional differentiation of hMSCs in one step without additional cytokine support. Both the hepatic biofunctions and transcriptome-based bioinformatics analysis indicated that TDN-miR122-hMSCs possessed the mature hepatocyte phenotype. In vivo study showed that intravenous transplantation of TDN-miR122-hMSCs was effective in rescuing ALF by improving liver functions and restoring injured livers with few side effects. It was worth noting that the collaboration of TDN as an antioxidant and TDN-miR122-hMSCs could well protect TDN-miR122-hMSCs activity, weaken its damage in ALF lesions, and improve its utilization. Collectively, the present study provides a new and simple strategy to induce the differentiation of hMSCs into functional hepatocyte-like cells based on TDN-miR122, which represents a feasible and alternative cell source for hepatic regeneration for alleviating ALF injury. Moreover, the TDNs-based nanoplatform provides extendable strategies for other DNA and RNA cargos delivery in various biomedical fields. Further large animal model explorations and understanding of the hepatic differentiation degree and biofunctions of TDN-miR122-hMSCs are needed for the confirmation of their potential in clinical translation.
Ethics approval and consent to participate
All the animal studies were performed following the protocol approved by the Institutional Animal Care and Use Committee of Sun Yat-Sen University.
CRediT authorship contribution statement
Hongyan Wei: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft. Fenfang Li: Methodology, Validation. Tiantian Xue: Methodology. Haixia Wang: Software, Supervision. Enguo Ju: Software, Supervision. Mingqiang Li: Conceptualization, Methodology, Visualization, Project administration, Resources, Supervision, Writing - review & editing. Yu Tao: Conceptualization, Methodology, Investigation, Validation, Visualization, Writing – review & editing, Funding acquisition, Project administration, Resources, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by the National Key Research and Development Program of China (2019YFA0111300), the Guangdong Provincial Pearl Talents Program (2019QN01Y131), the Thousand Talents Plan, and the Medical Science and Technology Research Fund of Guangdong Province (A2022112).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2023.04.024.
Contributor Information
Mingqiang Li, Email: limq567@mail.sysu.edu.cn.
Yu Tao, Email: taoy28@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wu H., Xia F., Zhang L., Fang C., Lee J., Gong L., Gao J., Ling D., Li F. A ROS-sensitive nanozyme-augmented photoacoustic nanoprobe for early diagnosis and therapy of acute liver failure. Adv. Mater. 2022;34(7) doi: 10.1002/adma.202108348. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez J., Acevedo J., Wiest R., Gustot T., Amoros A., Deulofeu C., Reverter E., Martinez J., Saliba F., Jalan R., Welzel T., Pavesi M., Hernandez-Tejero M., Gines P., Arroyo V. Bacterial and fungal infections in acute-on-chronic liver failure: prevalence, characteristics and impact on prognosis. Gut. 2018;67(10):1870–1880. doi: 10.1136/gutjnl-2017-314240. [DOI] [PubMed] [Google Scholar]
- 3.Kakabadze Z., Kakabadze A., Chakhunashvili D., Karalashvili L., Berishvili E., Sharma Y., Gupta S. Decellularized human placenta supports hepatic tissue and allows rescue in acute liver failure. Hepatology. 2018;67(5):1956–1969. doi: 10.1002/hep.29713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z., Li Y., Li W., Xiao C., Liu D., Dong C., Zhang M., Mäkilä E., Kemell M., Salonen J., Hirvonen J.T., Zhang H., Zhou D., Deng X., Santos H.A. Multifunctional nanohybrid based on porous silicon nanoparticles, gold nanoparticles, and acetalated dextran for liver regeneration and acute liver failure theranostics. Adv. Mater. 2018;30(24) doi: 10.1002/adma.201703393. [DOI] [PubMed] [Google Scholar]
- 5.Jin Y., Wang H., Yi K., Lv S., Hu H., Li M., Tao Y. Applications of nanobiomaterials in the therapy and imaging of acute liver failure. Nano-Micro Lett. 2021;13(1):25. doi: 10.1007/s40820-020-00550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolodziejczyk A.A., Federici S., Zmora N., Mohapatra G., Dori-Bachash M., Hornstein S., Leshem A., Reuveni D., Zigmond E., Tobar A., Salame T.M., Harmelin A., Shlomai A., Shapiro H., Amit I., Elinav E. Acute liver failure is regulated by MYC- and microbiome-dependent programs. Nat. Med. 2020;26(12):1899–1911. doi: 10.1038/s41591-020-1102-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., Chan H.F., Wang H., Shao D., Tao Y., Li M. Stem cell therapy and tissue engineering strategies using cell aggregates and decellularized scaffolds for the rescue of liver failure. J. Tissue Eng. 2021;12 doi: 10.1177/2041731420986711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J., Xu Y., Zhuo C., Shi R., Wang H., Hu Z., Chan H.F., Kim H.-W., Tao Y., Li M. Highly efficient fabrication of functional hepatocyte spheroids by a magnetic system for the rescue of acute liver failure. Biomaterials. 2023;294 doi: 10.1016/j.biomaterials.2023.122014. [DOI] [PubMed] [Google Scholar]
- 9.Weng J., Han X., Zeng F., Zhang Y., Feng L., Cai L., Liang K., Liu S., Li S., Fu G., Zeng M., Gao Y. Fiber scaffold bioartificial liver therapy relieves acute liver failure and extrahepatic organ injury in pigs. Theranostics. 2021;11(16):7620–7639. doi: 10.7150/thno.58515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W.-J., Zhu X.-J., Yuan T.-J., Wang Z.-Y., Bian Z.-Q., Jing H.-S., Shi X., Chen C.-Y., Fu G.-B., Huang W.-J., Shi Y.-P., Liu Q., Zeng M., Zhang H.-D., Wu H.-P., Yu W.-F., Zhai B., Yan H.-X. An extracorporeal bioartificial liver embedded with 3D-layered human liver progenitor-like cells relieves acute liver failure in pigs. Sci. Transl. Med. 2020;12(551) doi: 10.1126/scitranslmed.aba5146. eaba5146. [DOI] [PubMed] [Google Scholar]
- 11.Wang J., Huang D., Ren H., Shang L. Biomimic trained immunity-MSCs delivery microcarriers for acute liver failure regeneration. Small. 2022;18(36) doi: 10.1002/smll.202200858. [DOI] [PubMed] [Google Scholar]
- 12.Lee C.-W., Chen Y.-F., Wu H.-H., Lee O.K. Historical perspectives and advances in mesenchymal stem cell research for the treatment of liver diseases. Gastroenterology. 2018;154(1):46–56. doi: 10.1053/j.gastro.2017.09.049. [DOI] [PubMed] [Google Scholar]
- 13.Kim W., Gwon Y., Park S., Kim H., Kim J. Therapeutic strategies of three-dimensional stem cell spheroids and organoids for tissue repair and regeneration. Bioact. Mater. 2023;19:50–74. doi: 10.1016/j.bioactmat.2022.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alfaifi M., Eom Y.W., Newsome P.N., Baik S.K. Mesenchymal stromal cell therapy for liver diseases. J. Hepatol. 2018;68(6):1272–1285. doi: 10.1016/j.jhep.2018.01.030. [DOI] [PubMed] [Google Scholar]
- 15.Wirth K., Kizy S., Steer C. Liver regeneration in the acute liver failure patient. Clin. Liver Dis. 2018;22:269–287. doi: 10.1016/j.cld.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Chien Y., Chang Y.-L., Li H.-Y., Larsson M., Wu W.-W., Chien C.-S., Wang C.-Y., Chu P.-Y., Chen K.-H., Lo W.-L., Chiou S.-H., Lan Y.-T., Huo T.-I., Lee S.-D., Huang P.-I. Synergistic effects of carboxymethyl-hexanoyl chitosan, cationic polyurethane-short branch PEI in miR122 gene delivery: accelerated differentiation of iPSCs into mature hepatocyte-like cells and improved stem cell therapy in a hepatic failure model. Acta Biomater. 2015;13:228–244. doi: 10.1016/j.actbio.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 17.Davoodian N., Lotfi A.S., Soleimani M., Mowla S.J. MicroRNA-122 overexpression promotes hepatic differentiation of human adipose tissue-derived stem cells. J. Cell. Biochem. 2014;115(9):1582–1593. doi: 10.1002/jcb.24822. [DOI] [PubMed] [Google Scholar]
- 18.Xu H., He J.H., Xiao Z.D., Zhang Q.Q., Chen Y.Q., Zhou H., Qu L.H. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52(4):1431–1442. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 19.Deng X.G., Qiu R.L., Wu Y.H., Li Z.X., Xie P., Zhang J., Zhou J.J., Zeng L.X., Tang J., Maharjan A., Deng J.M. Overexpression of miR-122 promotes the hepatic differentiation and maturation of mouse ESCs through a miR-122/FoxA1/HNF4a-positive feedback loop. Liver Int. 2014;34(2):281–295. doi: 10.1111/liv.12239. [DOI] [PubMed] [Google Scholar]
- 20.Yang H., Han M., Li J., Ke H., Kong Y., Wang W., Wang L., Ma W., Qiu J., Wang X., Xin T., Liu H. Delivery of miRNAs through metal-organic framework nanoparticles for assisting neural stem cell therapy for ischemic stroke. ACS Nano. 2022;16(9):14503–14516. doi: 10.1021/acsnano.2c04886. [DOI] [PubMed] [Google Scholar]
- 21.Paunovska K., Loughrey D., Dahlman J.E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 2022;23(5):265–280. doi: 10.1038/s41576-021-00439-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huo D., Jiang X., Hu Y. Recent advances in nanostrategies capable of overcoming biological barriers for tumor management. Adv. Mater. 2020;32(27) doi: 10.1002/adma.201904337. [DOI] [PubMed] [Google Scholar]
- 23.Zhang T., Tian T., Zhou R., Li S., Ma W., Zhang Y., Liu N., Shi S., Li Q., Xie X., Ge Y., Liu M., Zhang Q., Lin S., Cai X., Lin Y. Design, fabrication and applications of tetrahedral DNA nanostructure-based multifunctional complexes in drug delivery and biomedical treatment. Nat. Protoc. 2020;15(8):2728–2757. doi: 10.1038/s41596-020-0355-z. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W., Callmann C.E., Meckes B., Mirkin C.A. Tumor-associated enzyme-activatable spherical nucleic acids. ACS Nano. 2022;16(7):10931–10942. doi: 10.1021/acsnano.2c03323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang D., Ge Z., Im H.J., England C.G., Ni D., Hou J., Zhang L., Kutyreff C.J., Yan Y., Liu Y., Cho S.Y., Engle J.W., Shi J., Huang P., Fan C., Yan H., Cai W. DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury. Nat. Biomed. Eng. 2018;2(11):865–877. doi: 10.1038/s41551-018-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Xiao L., Yan N., Li Y., Wang Y., Qin X., Zhao D., Liu M., Li N., Lin Y. The neuroprotective effect of microRNA-22-3p modified tetrahedral framework nucleic acids on damaged retinal neurons via TrkB/BDNF signaling pathway. Adv. Funct. Mater. 2021;31(36) [Google Scholar]
- 27.Su J., Wu F.B., Xia H.P., Wu Y.F., Liu S.Q. Accurate cancer cell identification and microRNA silencing induced therapy using tailored DNA tetrahedron nanostructures. Chem. Sci. 2020;11(1):80–86. doi: 10.1039/c9sc04823e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu L., Li P., Zhu J., Liao Z., Gao C., Li H., Yang Z., Zhao T., Chen W., Peng Y., Cao F., Ning C., Sui X., Guo Q., Lin Y., Liu S. Tetrahedral framework nucleic acids promote the biological functions and related mechanism of synovium-derived mesenchymal stem cells and show improved articular cartilage regeneration activity in situ. Bioact. Mater. 2022;9:411–427. doi: 10.1016/j.bioactmat.2021.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P., Ouyang Y., Sohn Y.S., Nechushtai R., Pikarsky E., Fan C., Willner I. pH- and miRNA-responsive DNA-tetrahedra/metal–organic framework conjugates: functional sense-and-treat carriers. ACS Nano. 2021;15(4):6645–6657. doi: 10.1021/acsnano.0c09996. [DOI] [PubMed] [Google Scholar]
- 30.Lee H., Lytton-Jean A.K.R., Chen Y., Love K.T., Park A.I., Karagiannis E.D., Sehgal A., Querbes W., Zurenko C.S., Jayaraman M., Peng C.G., Charisse K., Borodovsky A., Manoharan M., Donahoe J.S., Truelove J., Nahrendorf M., Langer R., Anderson D.G. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012;7(6):389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin X., Xiao L., Li N., Hou C., Li W., Li J., Yan N., Lin Y. Tetrahedral framework nucleic acids-based delivery of microRNA-155 inhibits choroidal neovascularization by regulating the polarization of macrophages. Bioact. Mater. 2022;14:134–144. doi: 10.1016/j.bioactmat.2021.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodman R.P., Schaap I.A.T., Tardin C.F., Erben C.M., Berry R.M., Schmidt C.F., Turberfield A.J. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310(5754):1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 33.Del Grosso E., Franco E., Prins L.J., Ricci F. Dissipative DNA nanotechnology. Nat. Chem. 2022;14(6):600–613. doi: 10.1038/s41557-022-00957-6. [DOI] [PubMed] [Google Scholar]
- 34.Zhang M., Zhang X., Tian T., Zhang Q., Wen Y., Zhu J., Xiao D., Cui W., Lin Y. Anti-inflammatory activity of curcumin-loaded tetrahedral framework nucleic acids on acute gouty arthritis. Bioact. Mater. 2022;8:368–380. doi: 10.1016/j.bioactmat.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., Yao Y., Wang Y., Xu J., Zhao D., Liu M., Shi S., Lin Y. Modulation of the crosstalk between schwann cells and macrophages for nerve regeneration: a therapeutic strategy based on a multifunctional tetrahedral framework nucleic acids system. Adv. Mater. 2022;34(46) doi: 10.1002/adma.202202513. [DOI] [PubMed] [Google Scholar]
- 36.Zhu J., Yang Y., Ma W., Wang Y., Chen L., Xiong H., Yin C., He Z., Fu W., Xu R., Lin Y. Antiepilepticus effects of tetrahedral framework nucleic acid via inhibition of gliosis-induced downregulation of glutamine synthetase and increased AMPAR internalization in the postsynaptic membrane. Nano Lett. 2022;22(6):2381–2390. doi: 10.1021/acs.nanolett.2c00025. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Q., Lin S., Shi S., Zhang T., Ma Q., Tian T., Zhou T., Cai X., Lin Y. Anti-inflammatory and antioxidative effects of tetrahedral DNA nanostructures via the modulation of macrophage responses. ACS Appl. Mater. Interfaces. 2018;10(4):3421–3430. doi: 10.1021/acsami.7b17928. [DOI] [PubMed] [Google Scholar]
- 38.Liu N., Zhang X., Li N., Zhou M., Zhang T., Li S., Cai X., Ji P., Lin Y. Tetrahedral framework nucleic acids promote corneal epithelial wound healing in vitro and in vivo. Small. 2019;15(31):1907–1917. doi: 10.1002/smll.201901907. [DOI] [PubMed] [Google Scholar]
- 39.Zhou M., Liu N.-X., Shi S.-R., Li Y., Zhang Q., Ma Q.-Q., Tian T., Ma W.-J., Cai X.-x., Lin Y.-F. Effect of tetrahedral DNA nanostructures on proliferation and osteo/odontogenic differentiation of dental pulp stem cells via activation of the notch signaling pathway. Nanomedicine. 2018;14(4):1227–1236. doi: 10.1016/j.nano.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Wei H., Zhao Z., Wang Y., Zou J., Lin Q., Duan Y. One-step self-assembly of multifunctional DNA nanohydrogels: an enhanced and harmless strategy for guiding combined antitumor therapy. ACS Appl. Mater. Interfaces. 2019;11(50):46479–46489. doi: 10.1021/acsami.9b15874. [DOI] [PubMed] [Google Scholar]
- 41.Zheng C., Zhang J., Chan H.F., Hu H., Lv S., Na N., Tao Y., Li M. Engineering nano-therapeutics to boost adoptive cell therapy for cancer treatment. Small Methods. 2021;5(5) doi: 10.1002/smtd.202001191. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Y., Gong Y., Chen X., Zhang Q., Zhang X., He Y., Pan L., Ni B., Yang F., Xu Y., Zhou L., Yang Y., Chen W. Injectable adhesive hemostatic gel with tumor acidity neutralizer and neutrophil extracellular traps lyase for enhancing adoptive NK cell therapy prevents post-resection recurrence of hepatocellular carcinoma. Biomaterials. 2022;284 doi: 10.1016/j.biomaterials.2022.121506. [DOI] [PubMed] [Google Scholar]
- 43.Wu X., Liu S., Zhu H., Ma Z., Dai X., Liu W. Scavenging ROS to alleviate acute liver injury by ZnO-NiO@COOH. Adv. Sci. 2022;9(11) doi: 10.1002/advs.202103982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao C., Li Z., Chen J., Su L., Wang J., Chen D.S., Ye J., Liao N., Yang H., Song J., Shi J. Site-specific biomimicry of antioxidative melanin formation and its application for acute liver injury therapy and imaging. Adv. Mater. 2021;33(34) doi: 10.1002/adma.202102391. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y., Shi S., Li B., Lan T., Yuan K., Yuan J., Zhou Y., Song J., Lv T., Shi Y., Xiang B., Tian T., Zhang T., Yang J., Lin Y. Therapeutic effects of self-assembled tetrahedral framework nucleic acids on liver regeneration in acute liver failure. ACS Appl. Mater. Interfaces. 2022;14(11):13136–13146. doi: 10.1021/acsami.2c02523. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., Li Y., Gao S., Yu X., Chen Y., Lin Y. Tetrahedral framework nucleic acids can alleviate taurocholate-induced severe acute pancreatitis and its subsequent multiorgan injury in mice. Nano Lett. 2022;22(4):1759–1768. doi: 10.1021/acs.nanolett.1c05003. [DOI] [PubMed] [Google Scholar]
- 47.Qiao Y., Xu Z., Yu Y., Hou S., Geng J., Xiao T., Liang Y., Dong Q., Mei Y., Wang B., Qiao H., Dai J., Suo G. Single cell derived spheres of umbilical cord mesenchymal stem cells enhance cell stemness properties, survival ability and therapeutic potential on liver failure. Biomaterials. 2020;227 doi: 10.1016/j.biomaterials.2019.119573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.