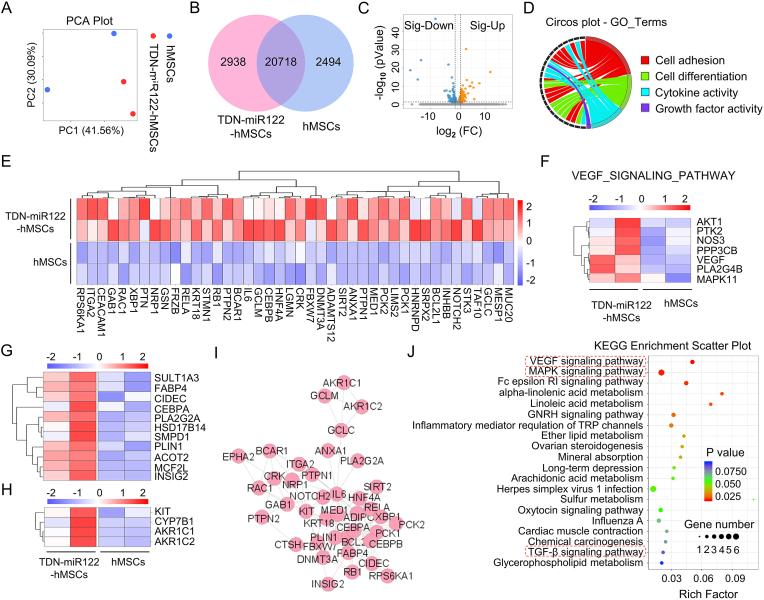
Fig. 4.
Transcriptome analysis reveals the underlying action mechanism of TDN-miR122 induced hMSCs-Hep. (A) Principal component analysis (PCA) was performed based on differentially expressed genes from TDN-miR122-hMSCs and hMSCs groups. Each data point corresponds to the PCA analysis of each sample. (B) Venn diagram of mRNAs differentially expressed in TDN-miR122-hMSCs groups and hMSCs groups. (C) A volcano plot showing the differences in gene expression between TDN-miR122-hMSCs groups and hMSCs groups. Blue points represent genes that were significantly reduced with a fold change >2, and red points represented significantly upregulated genes with a fold change >2. Gray points represents no significant difference. (D) Gene ontology (GO) enrichment analysis of the subordination between the representative differentially expressed genes and their enriched pathways. (E) Heatmap representing the expression of genes relating to specific liver functions (hepatocyte-related genes) in TDN-miR122-hMSCs groups and hMSCs groups, which further verified that hMSCs-Hep had the biological function of hepatocytes. (F) Heatmap representing the expression of genes relating to the VEGF signaling pathway in TDN-miR122-hMSCs groups and hMSCs groups. (G) Heatmap representing the expression of genes relating to the bile acid synthesis in TDN-miR122-hMSCs groups and hMSCs groups. (H) Heatmap representing the expression of genes relating to the fat and lipid metabolism in TDN-miR122-hMSCs groups and hMSCs groups. (I) PPI network consists of representative genes related to hepatic function and metabolism process markers, circles represent genes, and area of the circle represents the degree of genes. (J) Bubble charts show the KEGG pathway analysis results of differentially expressed genes (Top 20 KEGG pathways of differentially expressed genes).