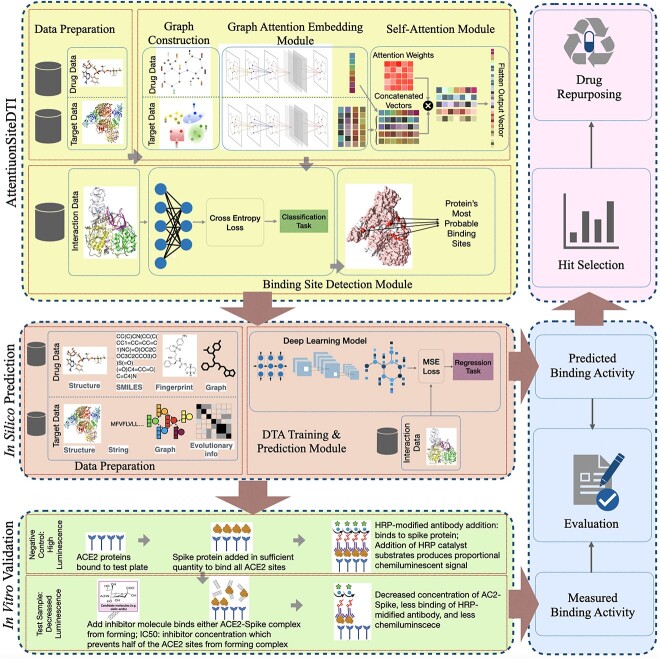
Figure 1.
Our proposed framework includes three main modules: (1) in-silico prediction that consists of a DL-based DTA prediction model augmented with AttentionSiteDTI for finding the most probable binding sites of proteins; (2) in-vitro validation, where we compare our computationally-predicted results with experimentally measured DTA values in laboratory to test and validate the practical potential of our proposed framework and (3) drug-repurposing module that utilizes our prediction model to identify hit compounds in prioritizing the most potent interactions for further in-vitro or ex-vivo verification in the laboratory.